MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction
- PMID: 19723330
- PMCID: PMC2745392
- DOI: 10.1186/1471-2105-10-274
MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction
Abstract
Background: Knowledge of subcellular localization of proteins is crucial to proteomics, drug target discovery and systems biology since localization and biological function are highly correlated. In recent years, numerous computational prediction methods have been developed. Nevertheless, there is still a need for prediction methods that show more robustness and higher accuracy.
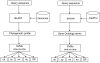
Results: We extended our previous MultiLoc predictor by incorporating phylogenetic profiles and Gene Ontology terms. Two different datasets were used for training the system, resulting in two versions of this high-accuracy prediction method. One version is specialized for globular proteins and predicts up to five localizations, whereas a second version covers all eleven main eukaryotic subcellular localizations. In a benchmark study with five localizations, MultiLoc2 performs considerably better than other methods for animal and plant proteins and comparably for fungal proteins. Furthermore, MultiLoc2 performs clearly better when using a second dataset that extends the benchmark study to all eleven main eukaryotic subcellular localizations.
Conclusion: MultiLoc2 is an extensive high-performance subcellular protein localization prediction system. By incorporating phylogenetic profiles and Gene Ontology terms MultiLoc2 yields higher accuracies compared to its previous version. Moreover, it outperforms other prediction systems in two benchmarks studies. MultiLoc2 is available as user-friendly and free web-service, available at: http://www-bs.informatik.uni-tuebingen.de/Services/MultiLoc2.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

