Signals and prepatterns: new insights into organ polarity in plants
- PMID: 19723761
- PMCID: PMC2751976
- DOI: 10.1101/gad.1819909
Signals and prepatterns: new insights into organ polarity in plants
Abstract
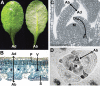
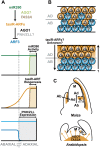
The flattening of leaves results from the interaction between upper (adaxial) and lower (abaxial) domains in the developing primordium. These domains are specified by conserved, overlapping genetic pathways involving several distinct transcription factor families and small regulatory RNAs. Polarity determinants employ a series of antagonistic interactions to produce mutually exclusive cell fates whose positioning is likely refined by signaling across the adaxial-abaxial boundary. Signaling candidates include a mobile small RNA-the first positional signal described in adaxial-abaxial polarity. Possible mechanisms to polarize the incipient primordium are discussed, including meristem-derived signaling and a model in which a polarized organogenic zone prepatterns the adaxial-abaxial axis.
Figures



References
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. - PubMed
-
- Allen E, Xie Z, Gustafson A, Carrington J. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. - PubMed
-
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40:1136–1141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
