Whole genome amplification and de novo assembly of single bacterial cells
- PMID: 19724646
- PMCID: PMC2731171
- DOI: 10.1371/journal.pone.0006864
Whole genome amplification and de novo assembly of single bacterial cells
Abstract
Background: Single-cell genome sequencing has the potential to allow the in-depth exploration of the vast genetic diversity found in uncultured microbes. We used the marine cyanobacterium Prochlorococcus as a model system for addressing important challenges facing high-throughput whole genome amplification (WGA) and complete genome sequencing of individual cells.
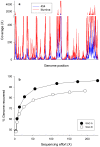
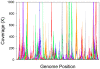
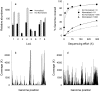
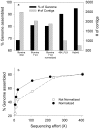
Methodology/principal findings: We describe a pipeline that enables single-cell WGA on hundreds of cells at a time while virtually eliminating non-target DNA from the reactions. We further developed a post-amplification normalization procedure that mitigates extreme variations in sequencing coverage associated with multiple displacement amplification (MDA), and demonstrated that the procedure increased sequencing efficiency and facilitated genome assembly. We report genome recovery as high as 99.6% with reference-guided assembly, and 95% with de novo assembly starting from a single cell. We also analyzed the impact of chimera formation during MDA on de novo assembly, and discuss strategies to minimize the presence of incorrectly joined regions in contigs.
Conclusions/significance: The methods describe in this paper will be useful for sequencing genomes of individual cells from a variety of samples.
Conflict of interest statement
Figures
References
-
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annual Review of Microbiology. 2003;57:369–394. - PubMed
-
- Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Applied Microbiology and Biotechnology. 2007;74:926–935. - PubMed
-
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11889–11894. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

