Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae
- PMID: 19726499
- PMCID: PMC2772707
- DOI: 10.1128/JVI.00677-09
Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae
Abstract
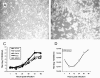
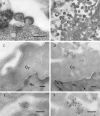
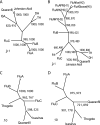
Arboviral infections are an important cause of emerging infections due to the movements of humans, animals, and hematophagous arthropods. Quaranfil virus (QRFV) is an unclassified arbovirus originally isolated from children with mild febrile illness in Quaranfil, Egypt, in 1953. It has subsequently been isolated in multiple geographic areas from ticks and birds. We used high-throughput sequencing to classify QRFV as a novel orthomyxovirus. The genome of this virus is comprised of multiple RNA segments; five were completely sequenced. Proteins with limited amino acid similarity to conserved domains in polymerase (PA, PB1, and PB2) and hemagglutinin (HA) genes from known orthomyxoviruses were predicted to be present in four of the segments. The fifth sequenced segment shared no detectable similarity to any protein and is of uncertain function. The end-terminal sequences of QRFV are conserved between segments and are different from those of the known orthomyxovirus genera. QRFV is known to cross-react serologically with two other unclassified viruses, Johnston Atoll virus (JAV) and Lake Chad virus (LKCV). The complete open reading frames of PB1 and HA were sequenced for JAV, while a fragment of PB1 of LKCV was identified by mass sequencing. QRFV and JAV PB1 and HA shared 80% and 70% amino acid identity to each other, respectively; the LKCV PB1 fragment shared 83% amino acid identity with the corresponding region of QRFV PB1. Based on phylogenetic analyses, virion ultrastructural features, and the unique end-terminal sequences identified, we propose that QRFV, JAV, and LKCV comprise a novel genus of the family Orthomyxoviridae.
Figures
References
-
- Al-Khalifa, M. S., F. M. Diab, and G. M. Khalil. 2007. Man-threatening viruses isolated from ticks in Saudi Arabia. Saudi Med. J. 28:1864-1867. - PubMed
-
- Austin, F. J. 1978. Johnston Atoll virus (Quaranfil group) from Ornithodoros capensis (Ixodoidea: Argasidae) infesting a gannet colony in New Zealand. Am. J. Trop. Med. Hyg. 27:1045-1048. - PubMed
-
- Beaty, B. J., C. H. Calisher, and R. E. Shope. 1989. Arboviruses, p. 797-855. In N. J. Schmidt and R. W. Emmons (ed.), Diagnostic procedures for viral, rickettsial, and chlamydial infections. American Public Health Association, Washington, DC.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

