Loss of the Brm-type SWI/SNF chromatin remodeling complex is a strong barrier to the Tat-independent transcriptional elongation of human immunodeficiency virus type 1 transcripts
- PMID: 19726504
- PMCID: PMC2772682
- DOI: 10.1128/JVI.00742-09
Loss of the Brm-type SWI/SNF chromatin remodeling complex is a strong barrier to the Tat-independent transcriptional elongation of human immunodeficiency virus type 1 transcripts
Abstract
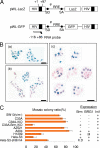
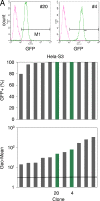
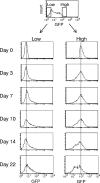
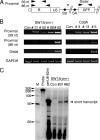
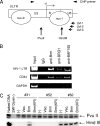
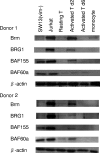
To elucidate the epigenetic regulation of Tat-independent human immunodeficiency virus (HIV) transcription following proviral integration, we constructed an HIV type 1 (HIV-1)-based replication-defective viral vector that expresses a reporter green fluorescent protein (GFP) product from its intact long terminal repeat (LTR). We transduced this construct into human tumor cell lines that were either deficient in or competent for the Brm-type SWI/SNF complex. One day after transduction, single cells that expressed GFP were sorted, and the GFP expression profiles originating from each of these clones were analyzed. Unlike clones of the SWI/SNF-competent cell line, which exhibited clear unimodal expression patterns in all cases, many clones originating from Brm-deficient cell lines either showed a broad-range distribution of GFP expression or were fully silenced. The resorting of GFP-negative populations of these isolated clones showed that GFP silencing is either reversible or irreversible depending upon the proviral integration sites. We further observed that even in these silenced clones, proviral gene transcription initiates to accumulate short transcripts of around 60 bases in length, but no elongation occurs. We found that this termination is caused by tightly closed nucleosome-1 (nuc-1) at the 5' LTR. Also, nuc-1 is remodeled by exogenous Brm in some integrants. From these results, we propose that Brm is required for the occasional transcriptional elongation of the HIV-1 provirus in the absence of Tat. Since the Brm-type SWI/SNF complex is expressed at marginal levels in resting CD4+ T cells and is drastically induced upon CD4+ T-cell activation, we speculate that it plays crucial roles in the early Tat-independent phase of HIV transcription in affected patients.
Figures

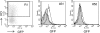

Similar articles
-
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription.Retrovirology. 2006 Aug 7;3:48. doi: 10.1186/1742-4690-3-48. Retrovirology. 2006. PMID: 16893449 Free PMC article.
-
Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter.EMBO J. 2006 Apr 19;25(8):1690-9. doi: 10.1038/sj.emboj.7601074. Epub 2006 Apr 6. EMBO J. 2006. PMID: 16601680 Free PMC article.
-
SNFing HIV transcription.Retrovirology. 2006 Aug 9;3:49. doi: 10.1186/1742-4690-3-49. Retrovirology. 2006. PMID: 16899112 Free PMC article.
-
Chromatin remodeling and modification during HIV-1 Tat-activated transcription.Curr HIV Res. 2003 Jul;1(3):343-62. doi: 10.2174/1570162033485186. Curr HIV Res. 2003. PMID: 15046258 Review.
-
Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription.Curr Pharm Des. 2017;23(28):4079-4090. doi: 10.2174/1381612823666170622104355. Curr Pharm Des. 2017. PMID: 28641539 Free PMC article. Review.
Cited by
-
Forging a Functional Cure for HIV: Transcription Regulators and Inhibitors.Viruses. 2022 Sep 7;14(9):1980. doi: 10.3390/v14091980. Viruses. 2022. PMID: 36146786 Free PMC article. Review.
-
Impact of chromatin on HIV-1 latency: a multi-dimensional perspective.Epigenetics Chromatin. 2025 Mar 8;18(1):9. doi: 10.1186/s13072-025-00573-x. Epigenetics Chromatin. 2025. PMID: 40055755 Free PMC article. Review.
-
Chemical probes targeting epigenetic proteins: Applications beyond oncology.Epigenetics. 2017 May 4;12(5):378-400. doi: 10.1080/15592294.2017.1279371. Epub 2017 Jan 12. Epigenetics. 2017. PMID: 28080202 Free PMC article. Review.
-
Curing HIV: Pharmacologic approaches to target HIV-1 latency.Annu Rev Pharmacol Toxicol. 2011;51:397-418. doi: 10.1146/annurev-pharmtox-010510-100237. Annu Rev Pharmacol Toxicol. 2011. PMID: 21210747 Free PMC article. Review.
-
Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway.J Biol Chem. 2010 Jul 16;285(29):21951-60. doi: 10.1074/jbc.M109.087783. Epub 2010 May 11. J Biol Chem. 2010. PMID: 20460684 Free PMC article.
References
-
- Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. - PMC - PubMed
-
- Adams, M., C. Wong, D. Wang, and J. Romeo. 1999. Limitation of Tat-associated transcriptional processivity in HIV-infected PBMC. Virology 257:397-405. - PubMed
-
- Arai, T., K. Matsumoto, K. Saitoh, M. Ui, T. Ito, M. Murakami, Y. Kanegae, I. Saito, F. L. Cosset, Y. Takeuchi, and H. Iba. 1998. A new system for stringent, high-titer vesicular stomatitis virus G protein-pseudotyped retrovirus vector induction by introduction of Cre recombinase into stable prepackaging cell lines. J. Virol. 72:1115-1121. - PMC - PubMed
-
- Arai, T., M. Takada, M. Ui, and H. Iba. 1999. Dose-dependent transduction of vesicular stomatitis virus G protein-pseudotyped retrovirus vector into human solid tumor cell lines and murine fibroblasts. Virology 260:109-115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

