SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering
- PMID: 19726574
- PMCID: PMC2773071
- DOI: 10.1104/pp.109.143941
SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering
Abstract
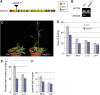
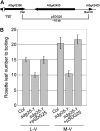
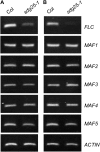
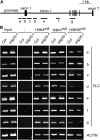
Covalent modifications of histone lysine residues by methylation play key roles in the regulation of chromatin structure and function. In contrast to H3K9 and H3K27 methylations that mark repressive states of transcription and are absent in some lower eukaryotes, H3K4 and H3K36 methylations are considered as active marks of transcription and are highly conserved in all eukaryotes from yeast (Saccharomyces cerevisiae) to Homo sapiens. Paradoxically, protein complexes catalyzing H3K4 and H3K36 methylations are less-extensively characterized in higher eukaryotes, particularly in plants. Arabidopsis (Arabidopsis thaliana) contains 12 SET DOMAIN GROUP (SDG) proteins phylogenetic classified to Trithorax Group (TrxG) and thus potentially involved in H3K4 and H3K36 methylations. So far only some genes of this family had been functionally characterized. Here we report on the genetic and molecular characterization of SDG25, a previously uncharacterized member of the Arabidopsis TrxG family. We show that the loss-of-function mutant sdg25-1 has an early flowering phenotype associated with suppression of FLOWERING LOCUS C (FLC) expression. Recombinant SDG25 proteins could methylate histone H3 from oligonucleosomes and mutant sdg25-1 plants showed weakly reduced levels of H3K36 dimethylation at FLC chromatin. Interestingly, sdg25-1 transcriptome shared a highly significant number of differentially expressed genes with that of sdg26-1, a previously characterized mutant exhibiting late-flowering phenotype and elevated FLC expression. Taken together, our results provide, to our knowledge, the first demonstration for a biological function of SDG25 and reveal additional layers of complexity of overlap and nonoverlap functions of the TrxG family genes in Arabidopsis.
Figures


References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 15 627–637 - PubMed
-
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29 4319–4333 - PMC - PubMed
-
- Baurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125 655–664 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

