Two-dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside-RNA hairpin loop interactions
- PMID: 19726586
- PMCID: PMC2761267
- DOI: 10.1093/nar/gkp594
Two-dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside-RNA hairpin loop interactions
Abstract
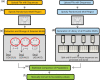
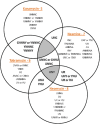
Herein, we report the identification of RNA hairpin loops that bind derivatives of kanamycin A, tobramycin, neamine, and neomycin B via two-dimensional combinatorial screening, a method that screens chemical and RNA spaces simultaneously. An arrayed aminoglycoside library was probed for binding to a 6-nucleotide RNA hairpin loop library (4096 members). Members of the loop library that bound each aminoglycoside were excised from the array, amplified and sequenced. Sequences were analyzed with our newly developed RNA Privileged Space Predictor (RNA-PSP) program, which analyzes selected sequences to identify statistically significant trends. RNA-PSP identified the following unique trends: 5'UNNNC3' loops for the kanamycin A derivative (where N is any nucleotide); 5'UNNC3' loops for the tobramycin derivative; 5'UNC3' loops for the neamine derivative; and 5'UNNG3' loops for the neomycin B derivative. The affinities and selectivities of a subset of the ligand-hairpin loop interactions were determined. The selected interactions have K(d) values ranging from 10 nM to 605 nM. Selectivities ranged from 0.4 to >200-fold. Interestingly, the results from RNA-PSP are able to qualitatively predict specificity based on overlap between the RNA sequences selected for the ligands. These studies expand the information available on small molecule-RNA motif interactions, which could be useful to design ligands targeting RNA.
Figures




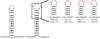
Similar articles
-
Defining RNA motif-aminoglycoside interactions via two-dimensional combinatorial screening and structure-activity relationships through sequencing.Bioorg Med Chem. 2013 Oct 15;21(20):6132-8. doi: 10.1016/j.bmc.2013.04.072. Epub 2013 May 7. Bioorg Med Chem. 2013. PMID: 23719281 Free PMC article.
-
Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners.J Am Chem Soc. 2008 Aug 20;130(33):11185-94. doi: 10.1021/ja803234t. Epub 2008 Jul 25. J Am Chem Soc. 2008. PMID: 18652457
-
Two-dimensional combinatorial screening of a bacterial rRNA A-site-like motif library: defining privileged asymmetric internal loops that bind aminoglycosides.Biochemistry. 2010 Mar 9;49(9):1833-42. doi: 10.1021/bi901998m. Biochemistry. 2010. PMID: 20108982 Free PMC article.
-
Two-dimensional combinatorial screening identifies specific 6'-acylated kanamycin A- and 6'-acylated neamine-RNA hairpin interactions.Biochemistry. 2008 Dec 2;47(48):12670-9. doi: 10.1021/bi8012615. Biochemistry. 2008. PMID: 18991404 Free PMC article.
-
Thermodynamics of aminoglycoside-rRNA recognition.Biopolymers. 2003 Sep;70(1):58-79. doi: 10.1002/bip.10411. Biopolymers. 2003. PMID: 12925993 Review.
Cited by
-
Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays.J Am Chem Soc. 2014 Jun 11;136(23):8402-10. doi: 10.1021/ja502754f. Epub 2014 May 28. J Am Chem Soc. 2014. PMID: 24820959 Free PMC article.
-
Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations.Nat Commun. 2012;3:1125. doi: 10.1038/ncomms2119. Nat Commun. 2012. PMID: 23047683 Free PMC article.
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy.Nat Commun. 2023 Oct 3;14(1):6166. doi: 10.1038/s41467-023-41799-1. Nat Commun. 2023. PMID: 37789015 Free PMC article.
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy.bioRxiv [Preprint]. 2023 Jun 15:2023.06.15.545153. doi: 10.1101/2023.06.15.545153. bioRxiv. 2023. Update in: Nat Commun. 2023 Oct 3;14(1):6166. doi: 10.1038/s41467-023-41799-1. PMID: 37397986 Free PMC article. Updated. Preprint.
-
Defining RNA motif-aminoglycoside interactions via two-dimensional combinatorial screening and structure-activity relationships through sequencing.Bioorg Med Chem. 2013 Oct 15;21(20):6132-8. doi: 10.1016/j.bmc.2013.04.072. Epub 2013 May 7. Bioorg Med Chem. 2013. PMID: 23719281 Free PMC article.
References
-
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. - PubMed
-
- Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 2007;120:1833–1840. - PubMed
-
- Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. - PubMed
-
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

