Design and validation of real-time reverse transcription-PCR assays for detection of pandemic (H1N1) 2009 virus
- PMID: 19726603
- PMCID: PMC2772599
- DOI: 10.1128/JCM.01103-09
Design and validation of real-time reverse transcription-PCR assays for detection of pandemic (H1N1) 2009 virus
Abstract
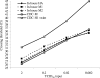
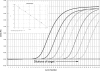
Tracking novel influenza viruses which have the potential to cause pandemics, such as the pandemic (H1N1) 2009 virus, is a public health priority. Pandemic (H1N1) 2009 virus was first identified in Mexico in April 2009 and spread worldwide over a short period of time. Well-validated diagnostic tools that are rapid, sensitive, and specific for the detection and tracking of this virus are needed. Three real-time reverse transcription PCR (RT-PCR) assays for the amplification and detection of pandemic (H1N1) 2009 virus were developed, and their performance characteristics were compared with those of other published diagnostic assays. Thirty-nine samples confirmed to be positive for pandemic (H1N1) 2009 virus from Alberta, Canada, and six additional samples that were positive for influenza A virus but that were not typeable by using published seasonal influenza H1/H3 virus assays were available for this validation. Amplification and direct sequencing of the products was considered the "gold standard" for case identification. The new assays were sensitive and able to reproducibly detect virus in a 10(-6) dilution of 4 x 10(6) 50% tissue culture infective doses/ml when 5 microl was used as the template. They showed 100% specificity and did not cross-react with other respiratory viruses or seasonal influenza A virus subtypes. The coefficient of variation in crossing cycle threshold values for the detection of different template concentrations of pandemic (H1N1) 2009 virus was < or =3.13%, showing good reproducibility. The assays had a wide dynamic range for the detection of pandemic (H1N1) 2009 virus and utilized testing platforms appropriate for high diagnostic throughput with rapid turnaround times. We developed and validated these real-time PCR procedures with the goal that they will be useful for diagnosis and surveillance of pandemic (H1N1) 2009 virus. These findings will contribute to the informed management of this novel virus.
Figures
References
-
- Anonymous. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. - PubMed
-
- Centers for Disease Control and Prevention. 2009. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467-470. - PubMed
-
- Centers for Disease Control and Prevention. 2009. Update: novel influenza A (H1N1) virus infections—worldwide, May 6, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:453-458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

