Single nucleotide polymorphisms on the road to strain differentiation in Mycobacterium ulcerans
- PMID: 19726608
- PMCID: PMC2772594
- DOI: 10.1128/JCM.00761-09
Single nucleotide polymorphisms on the road to strain differentiation in Mycobacterium ulcerans
Abstract
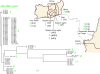
The genomic fine-typing of strains of Mycobacterium ulcerans, the causative agent of the emerging human disease Buruli ulcer, is difficult due to the clonal population structure of geographical lineages. Although large sequence polymorphisms (LSPs) resulted in the clustering of patient isolates originating from across the globe, differentiation of strains within continents using conventional typing methods is very limited. In this study, we analyzed M. ulcerans LSP haplotype-specific insertion sequence elements among 83 M. ulcerans strains and identified single nucleotide polymorphisms (SNPs) that differentiate between regional strains. This is the first genetic discrimination based on SNPs of M. ulcerans strains from African countries where Buruli ulcer is endemic, resulting in the highest geographic resolution of genotyping so far. The findings support the concept of genome-wide SNP analyses as tools to study the epidemiology and evolution of M. ulcerans at a local level.
Figures
References
-
- Ablordey, A., P. A. Fonteyne, P. Stragier, P. Vandamme, and F. Portaels. 2007. Identification of a new variable number tandem repeat locus in Mycobacterium ulcerans for potential strain discrimination among African isolates. Clin. Microbiol. Infect. 13:734-736. - PubMed
-
- Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

