An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator
- PMID: 19726650
- PMCID: PMC2757269
- DOI: 10.1523/JNEUROSCI.2133-09.2009
An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator
Abstract
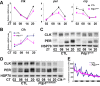
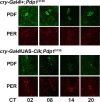
The Drosophila PAR domain protein 1 (Pdp1) gene encodes a transcription factor with multiple functions. One isoform, PDP1epsilon, was proposed to be an essential activator of the core clock gene, Clock (Clk). However, a central clock function for PDP1epsilon was recently disputed, and genetic analysis has been difficult due to developmental lethality of Pdp1-null mutants. Here we report the discovery of a mutation that specifically disrupts the Pdp1epsilon isoform. Homozygous Pdp1epsilon mutants are viable and exhibit arrhythmic circadian behavior in constant darkness and also in the presence of light:dark cycles. Importantly, the mutants show diminished expression of CLK and PERIOD (PER) in the central clock cells. In addition, expression of PDF (pigment-dispersing factor) is reduced in a subset of the central clock cells. Loss of Pdp1epsilon also alters the phosphorylation status of the CLK protein and disrupts cyclic expression of a per-luciferase reporter in peripheral clocks under free-running conditions. Transgenic expression of PDP1epsilon in clock neurons of Pdp1epsilon mutants can restore rhythmic circadian behavior. However, transgenic expression of CLK in these mutants rescues the expression of PER in the central clock, but fails to restore behavioral rhythms, suggesting that PDP1epsilon has effects outside the core molecular clock. Together, these data support a model in which PDP1epsilon functions in the central circadian oscillator as well as in the output pathway.
Figures

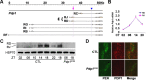
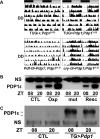
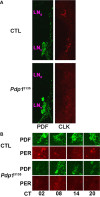
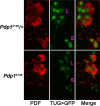
Similar articles
-
PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms.J Neurosci. 2007 Mar 7;27(10):2539-47. doi: 10.1523/JNEUROSCI.4870-06.2007. J Neurosci. 2007. PMID: 17344391 Free PMC article.
-
Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster.Biochem Biophys Res Commun. 2007 Dec 14;364(2):294-300. doi: 10.1016/j.bbrc.2007.10.009. Epub 2007 Oct 12. Biochem Biophys Res Commun. 2007. PMID: 17950247
-
The circadian output gene takeout is regulated by Pdp1epsilon.Proc Natl Acad Sci U S A. 2010 Feb 9;107(6):2544-9. doi: 10.1073/pnas.0906422107. Epub 2010 Jan 21. Proc Natl Acad Sci U S A. 2010. PMID: 20133786 Free PMC article.
-
Transcriptional feedback loop regulation, function, and ontogeny in Drosophila.Cold Spring Harb Symp Quant Biol. 2007;72:437-44. doi: 10.1101/sqb.2007.72.009. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419302 Free PMC article. Review.
-
Roles of peripheral clocks: lessons from the fly.FEBS Lett. 2022 Feb;596(3):263-293. doi: 10.1002/1873-3468.14251. Epub 2021 Dec 16. FEBS Lett. 2022. PMID: 34862983 Free PMC article. Review.
Cited by
-
Regulation of gustatory physiology and appetitive behavior by the Drosophila circadian clock.Curr Biol. 2010 Feb 23;20(4):300-9. doi: 10.1016/j.cub.2009.12.055. Epub 2010 Feb 11. Curr Biol. 2010. PMID: 20153192 Free PMC article.
-
Fluorescent Reporters for Studying Circadian Rhythms in Drosophila melanogaster.Methods Mol Biol. 2022;2482:353-371. doi: 10.1007/978-1-0716-2249-0_24. Methods Mol Biol. 2022. PMID: 35610439
-
Circadian disruption of memory consolidation in Drosophila.Front Syst Neurosci. 2023 Mar 22;17:1129152. doi: 10.3389/fnsys.2023.1129152. eCollection 2023. Front Syst Neurosci. 2023. PMID: 37034015 Free PMC article.
-
A new promoter element associated with daily time keeping in Drosophila.Nucleic Acids Res. 2017 Jun 20;45(11):6459-6470. doi: 10.1093/nar/gkx268. Nucleic Acids Res. 2017. PMID: 28407113 Free PMC article.
-
Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing.Elife. 2019 Nov 8;8:e50063. doi: 10.7554/eLife.50063. Elife. 2019. PMID: 31702555 Free PMC article.
References
-
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. - PubMed
-
- Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
