MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib
- PMID: 19726678
- PMCID: PMC2797273
- DOI: 10.1074/jbc.M109.016774
MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib
Abstract
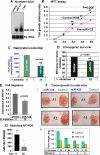
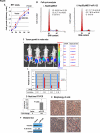
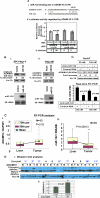
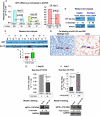
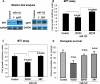
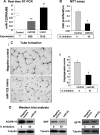
MicroRNAs are negative regulators of protein coding genes. The liver-specific microRNA-122 (miR-122) is frequently suppressed in primary hepatocellular carcinomas (HCCs). In situ hybridization demonstrated that miR-122 is abundantly expressed in hepatocytes but barely detectable in primary human HCCs. Ectopic expression of miR-122 in nonexpressing HepG2, Hep3B, and SK-Hep-1 cells reversed their tumorigenic properties such as growth, replication potential, clonogenic survival, anchorage-independent growth, migration, invasion, and tumor formation in nude mice. Further, miR-122-expressing HCC cells retained an epithelial phenotype that correlated with reduced Vimentin expression. ADAM10 (a distintegrin and metalloprotease family 10), serum response factor (SRF), and insulin-like growth factor 1 receptor (Igf1R) that promote tumorigenesis were validated as targets of miR-122 and were repressed by the microRNA. Conversely, depletion of the endogenous miR-122 in Huh-7 cells facilitated their tumorigenic properties with concomitant up-regulation of these targets. Expression of SRF or Igf1R partially reversed tumor suppressor function of miR-122. Further, miR-122 impeded angiogenic properties of endothelial cells in vitro. Notably, ADAM10, SRF, and Igf1R were up-regulated in primary human HCCs compared with the matching liver tissue. Co-labeling studies demonstrated exclusive localization of miR-122 in the benign livers, whereas SRF predominantly expressed in HCC. More importantly, growth and clonogenic survival of miR-122-expressing HCC cells were significantly reduced upon treatment with sorafenib, a multi-kinase inhibitor clinically effective against HCC. Collectively, these results suggest that the loss of multifunctional miR-122 contributes to the malignant phenotype of HCC cells, and miR-122 mimetic alone or in combination with anticancer drugs can be a promising therapeutic regimen against liver cancer.
Figures


References
-
- El-Serag H. B., Rudolph K. L. (2007) Gastroenterology 132, 2557–2576 - PubMed
-
- Visone R., Petrocca F., Croce C. M. (2008) Gastroenterology 135, 1866–1869 - PubMed
-
- Braconi C., Patel T. (2008) Hepatology 47, 1807–1809 - PubMed
-
- Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

