Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase
- PMID: 19726683
- PMCID: PMC2781579
- DOI: 10.1074/jbc.M109.046391
Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase
Abstract
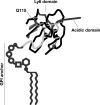
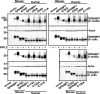
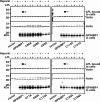
GPIHBP1, a glycosylphosphatidylinositol-anchored endothelial cell protein of the lymphocyte antigen 6 (Ly6) family, binds lipoprotein lipase (LPL) avidly and is required for the lipolytic processing of triglyceride-rich lipoproteins. GPIHBP1 contains two key structural motifs, an acidic domain and an Ly6 motif (a three-fingered domain specified by 10 cysteines). The acidic domain is required for LPL binding, but the importance of the Ly6 domain is less clear. To explore that issue, we transfected cells with a wild-type GPIHBP1 expression vector or mutant GPIHBP1 vectors in which specific cysteines in the Ly6 domain were changed to alanine. The mutant GPIHBP1 proteins reached the cell surface, as judged by antibody binding studies and by the ability of a phosphatidylinositol-specific phospholipase C to release these proteins from the cell surface. However, cells expressing the cysteine mutants could not bind LPL. The acidic domain of the cysteine mutants appeared to remain accessible, as judged by binding studies with an antibody against the acidic domain. We also developed a cell-free assay of LPL binding. We created a rat monoclonal antibody against the carboxyl terminus of mouse GPIHBP1 and used that antibody to coat agarose beads. We then tested the ability of soluble forms of GPIHBP1 that had been immobilized on monoclonal antibody-coated beads to bind LPL. In this assay, wild-type soluble GPIHBP1 bound LPL avidly, but the cysteine mutants did not. Thus, our studies suggest that a structurally intact Ly6 domain (in addition to the acidic domain) is essential for LPL binding.
Figures


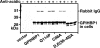

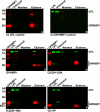
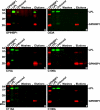
References
-
- Ioka R. X., Kang M. J., Kamiyama S., Kim D. H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., Sasano H., Takahashi S., Sakai J., Fujino T., Yamamoto T. T. (2003) J. Biol. Chem. 278, 7344–7349 - PubMed
-
- Hata A., Ridinger D. N., Sutherland S., Emi M., Shuhua Z., Myers R. L., Ren K., Cheng T., Inoue I., Wilson D. E., Iverius P. H., Lalouel J. M. (1993) J. Biol. Chem. 268, 8447–8457 - PubMed
-
- Ma Y., Henderson H. E., Liu M. S., Zhang H., Forsythe I. J., Clarke-Lewis I., Hayden M. R., Brunzell J. D. (1994) J. Lipid Res. 35, 2049–2059 - PubMed
-
- Sendak R. A., Bensadoun A. (1998) J. Lipid Res. 39, 1310–1315 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

