A sulfilimine bond identified in collagen IV
- PMID: 19729652
- PMCID: PMC2876822
- DOI: 10.1126/science.1176811
A sulfilimine bond identified in collagen IV
Abstract
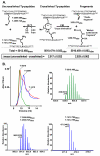
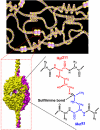
Collagen IV networks are ancient proteins of basement membranes that underlie epithelia in metazoa from sponge to human. The networks provide structural integrity to tissues and serve as ligands for integrin cell-surface receptors. They are assembled by oligomerization of triple-helical protomers and are covalently crosslinked, a key reinforcement that stabilizes networks. We used Fourier-transform ion cyclotron resonance mass spectrometry and nuclear magnetic resonance spectroscopy to show that a sulfilimine bond (-S=N-) crosslinks hydroxylysine-211 and methionine-93 of adjoining protomers, a bond not previously found in biomolecules. This bond, the nitrogen analog of a sulfoxide, appears to have arisen at the divergence of sponge and cnidaria, an adaptation of the extracellular matrix in response to mechanical stress in metazoan evolution.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

