Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors
- PMID: 19730695
- PMCID: PMC2731224
- DOI: 10.1371/journal.ppat.1000569
Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors
Abstract
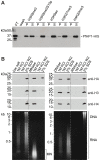
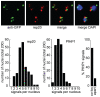
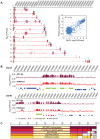
Epigenetic processes are the main conductors of phenotypic variation in eukaryotes. The malaria parasite Plasmodium falciparum employs antigenic variation of the major surface antigen PfEMP1, encoded by 60 var genes, to evade acquired immune responses. Antigenic variation of PfEMP1 occurs through in situ switches in mono-allelic var gene transcription, which is PfSIR2-dependent and associated with the presence of repressive H3K9me3 marks at silenced loci. Here, we show that P. falciparum heterochromatin protein 1 (PfHP1) binds specifically to H3K9me3 but not to other repressive histone methyl marks. Based on nuclear fractionation and detailed immuno-localization assays, PfHP1 constitutes a major component of heterochromatin in perinuclear chromosome end clusters. High-resolution genome-wide chromatin immuno-precipitation demonstrates the striking association of PfHP1 with virulence gene arrays in subtelomeric and chromosome-internal islands and a high correlation with previously mapped H3K9me3 marks. These include not only var genes, but also the majority of P. falciparum lineage-specific gene families coding for exported proteins involved in host-parasite interactions. In addition, we identified a number of PfHP1-bound genes that were not enriched in H3K9me3, many of which code for proteins expressed during invasion or at different life cycle stages. Interestingly, PfHP1 is absent from centromeric regions, implying important differences in centromere biology between P. falciparum and its human host. Over-expression of PfHP1 results in an enhancement of variegated expression and highlights the presence of well-defined heterochromatic boundaries. In summary, we identify PfHP1 as a major effector of virulence gene silencing and phenotypic variation. Our results are instrumental for our understanding of this widely used survival strategy in unicellular pathogens.
Conflict of interest statement
The authors have declared that no competing interest exists
Figures



References
-
- Pongponratn E, Riganti M, Punpoowong B, Aikawa M. Microvascular sequestration of parasitized erythrocytes in human falciparum malaria: a pathological study. Am J Trop Med Hyg. 1991;44:168–175. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

