Overview: generation of gene knockout mice
- PMID: 19731224
- PMCID: PMC2782548
- DOI: 10.1002/0471143030.cb1912s44
Overview: generation of gene knockout mice
Abstract
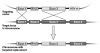
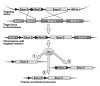
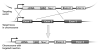
The technique of gene targeting allows for the introduction of engineered genetic mutations into a mouse at a determined genomic locus. The process of generating mouse models with targeted mutations was developed through both the discovery of homologous recombination and the isolation of murine embryonic stem cells (ES cells). Homologous recombination is a DNA repair mechanism that is employed in gene targeting to insert a designed mutation into the homologous genetic locus. Targeted homologous recombination can be performed in murine ES cells through electroporation of a targeting construct. These ES cells are totipotent and, when injected into a mouse blastocyst, they can differentiate into all cell types of a chimeric mouse. A chimeric mouse harboring cells derived from the targeted ES cell clone can then generate a whole mouse containing the desired targeted mutation. The initial step for the generation of a mouse with a targeted mutation is the construction of an efficient targeting vector that will be introduced into the ES cells.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

