Replication cycle of chikungunya: a re-emerging arbovirus
- PMID: 19732931
- PMCID: PMC2915564
- DOI: 10.1016/j.virol.2009.07.024
Replication cycle of chikungunya: a re-emerging arbovirus
Abstract
Arboviruses (or arthropod-borne viruses), represent a threat for the new century. The 2005-2006 year unprecedented epidemics of chikungunya virus (CHIKV) in the French Reunion Island in the Indian Ocean, followed by several outbreaks in other parts of the world such as India, have attracted the attention of clinicians, scientists, and state authorities about the risks linked to this re-emerging mosquito-borne virus. CHIKV, which belongs to the Alphaviruses genus, was not previously regarded as a highly pathogenic arbovirus. However, this opinion was challenged by the death of several CHIKV-infected persons in Reunion Island. The epidemic episode began in December 2005 and four months later the seroprevalence survey report indicated that 236,000 persons, more than 30% of Reunion Island population, had been infected with CHIKV, among which 0.4-0.5% of cases were fatal. Since the epidemic peak, the infection case number has continued to increase to almost 40% of the population, with a total of more than 250 fatalities. Although information available on CHIKV is growing quite rapidly, we are still far from understanding the strategies required for the ecologic success of this virus, virus replication, its interactions with its vertebrate hosts and arthropod vectors, and its genetic evolution. In this paper, we summarize the current knowledge of CHIKV genomic organization, cell tropism, and the virus replication cycle, and evaluate the possibility to predict its future evolution. Such understanding may be applied in order to anticipate future epidemics and reduce the incidence by development and application of, for example, vaccination and antiviral therapy.
Figures


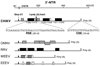

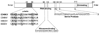



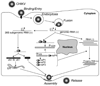
References
-
- Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 2007;88(Pt 7):1967–1976. - PubMed
-
- Briant L, Robert-Hebmann V, Sivan V, Brunet A, Pouyssegur J, Devaux C. Involvement of extracellular signal-regulated kinase module in HIV-mediated CD4 signals controlling activation of nuclear factor-kappa B and AP-1 transcription factors. J. Immunol. 1998;160(4):1875–1885. - PubMed
-
- Brighton SW. Chloroquine phosphate treatment of chronic chikungunya arthritis. An open pilot study. S. Afr. Med. J. 1984;66(6):217–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

