Identification of amino acids in the N-terminal domain of atypical methanogenic-type Seryl-tRNA synthetase critical for tRNA recognition
- PMID: 19734148
- PMCID: PMC2781618
- DOI: 10.1074/jbc.M109.044099
Identification of amino acids in the N-terminal domain of atypical methanogenic-type Seryl-tRNA synthetase critical for tRNA recognition
Abstract
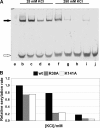
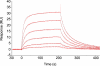
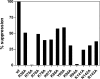
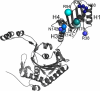
Seryl-tRNA synthetase (SerRS) from methanogenic archaeon Methanosarcina barkeri, contains an idiosyncratic N-terminal domain, composed of an antiparallel beta-sheet capped by a helical bundle, connected to the catalytic core by a short linker peptide. It is very different from the coiled-coil tRNA binding domain in bacterial-type SerRS. Because the crystal structure of the methanogenic-type SerRSxtRNA complex has not been obtained, a docking model was produced, which indicated that highly conserved helices H2 and H3 of the N-terminal domain may be important for recognition of the extra arm of tRNA(Ser). Based on structural information and the docking model, we have mutated various positions within the N-terminal region and probed their involvement in tRNA binding and serylation. Total loss of activity and inability of the R76A variant to form the complex with cognate tRNA identifies Arg(76) located in helix H2 as a crucial tRNA-interacting residue. Alteration of Lys(79) positioned in helix H2 and Arg(94) in the loop between helix H2 and beta-strand A4 have a pronounced effect on SerRSxtRNA(Ser) complex formation and dissociation constants (K(D)) determined by surface plasmon resonance. The replacement of residues Arg(38) (located in the loop between helix H1 and beta-strand A2), Lys(141) and Asn(142) (from H3), and Arg(143) (between H3 and H4) moderately affect both the serylation activity and the K(D) values. Furthermore, we have obtained a striking correlation between these results and in vivo effects of these mutations by quantifying the efficiency of suppression of bacterial amber mutations, after coexpression of the genes for M. barkeri suppressor tRNA(Ser) and a set of mMbSerRS variants in Escherichia coli.
Figures



Similar articles
-
Recognition between tRNASer and archaeal seryl-tRNA synthetases monitored by suppression of bacterial amber mutations.FEMS Microbiol Lett. 2009 May;294(1):111-8. doi: 10.1111/j.1574-6968.2009.01560.x. Epub 2008 Mar 20. FEMS Microbiol Lett. 2009. PMID: 19309487
-
Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.RNA Biol. 2008 Jul-Sep;5(3):169-77. doi: 10.4161/rna.5.3.6876. Epub 2008 Jul 28. RNA Biol. 2008. PMID: 18818520
-
Structural flexibility of the methanogenic-type seryl-tRNA synthetase active site and its implication for specific substrate recognition.FEBS J. 2008 Jun;275(11):2831-44. doi: 10.1111/j.1742-4658.2008.06423.x. Epub 2008 Apr 18. FEBS J. 2008. PMID: 18422966
-
Structure, function and evolution of seryl-tRNA synthetases: implications for the evolution of aminoacyl-tRNA synthetases and the genetic code.J Mol Evol. 1995 May;40(5):519-30. doi: 10.1007/BF00166620. J Mol Evol. 1995. PMID: 7540217 Review.
-
Escherichia coli seryl-tRNA synthetase: the structure of a class 2 aminoacyl-tRNA synthetase.Biochim Biophys Acta. 1991 Jul 23;1089(3):287-98. doi: 10.1016/0167-4781(91)90168-l. Biochim Biophys Acta. 1991. PMID: 1859832 Review. No abstract available.
Cited by
-
New aminoacyl-tRNA synthetase-like protein in insecta with an essential mitochondrial function.J Biol Chem. 2010 Dec 3;285(49):38157-66. doi: 10.1074/jbc.M110.167486. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870726 Free PMC article.
-
Biased gene transfer in microbial evolution.Nat Rev Microbiol. 2011 Jun 13;9(7):543-55. doi: 10.1038/nrmicro2593. Nat Rev Microbiol. 2011. PMID: 21666709 Review.
-
Insights into substrate promiscuity of human seryl-tRNA synthetase.RNA. 2017 Nov;23(11):1685-1699. doi: 10.1261/rna.061069.117. Epub 2017 Aug 14. RNA. 2017. PMID: 28808125 Free PMC article.
-
SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis.Nucleic Acids Res. 2015 Dec 2;43(21):10534-45. doi: 10.1093/nar/gkv996. Epub 2015 Oct 3. Nucleic Acids Res. 2015. PMID: 26433229 Free PMC article.
-
Emergence and evolution.Top Curr Chem. 2014;344:43-87. doi: 10.1007/128_2013_423. Top Curr Chem. 2014. PMID: 23478877 Free PMC article. Review.
References
-
- Ibba M., Soll D. (2000) Annu. Rev. Biochem. 69, 617–650 - PubMed
-
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Nature 347, 203–206 - PubMed
-
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. (1990) Nature 347, 249–255 - PubMed
-
- Ibba M., Morgan S., Curnow A. W., Pridmore D. R., Vothknecht U. C., Gardner W., Lin W., Woese C. R., Söll D. (1997) Science 278, 1119–1122 - PubMed
-
- Zhang C. M., Perona J. J., Ryu K., Francklyn C., Hou Y. M. (2006) J. Mol. Biol. 361, 300–311 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

