Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol
- PMID: 19734340
- PMCID: PMC2772450
- DOI: 10.1128/AEM.00670-09
Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol
Abstract
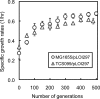
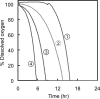
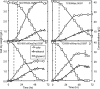
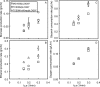
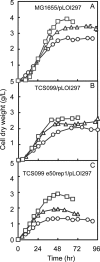
Based on elementary mode analysis, an Escherichia coli strain was designed for efficient conversion of glycerol to ethanol. By using nine gene knockout mutations, the functional space of the central metabolism of E. coli was reduced from over 15,000 possible pathways to a total of 28 glycerol-utilizing pathways that support cell function. Among these pathways are eight aerobic and eight anaerobic pathways that do not support cell growth but convert glycerol into ethanol with a theoretical yield of 0.50 g ethanol/g glycerol. The remaining 12 pathways aerobically coproduce biomass and ethanol from glycerol. The optimal ethanol production depends on the oxygen availability that regulates the two competing pathways for biomass and ethanol production. The coupling between cell growth and ethanol production enabled metabolic evolution of the designed strain through serial dilution that resulted in strains with improved ethanol yields and productivities. In defined medium, the evolved strain can convert 40 g/liter of glycerol to ethanol in 48 h with 90% of the theoretical ethanol yield. The performance of the designed strain is predicted by the property space of remaining elementary modes.
Figures






Similar articles
-
Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses.Appl Environ Microbiol. 2008 Jun;74(12):3634-43. doi: 10.1128/AEM.02708-07. Epub 2008 Apr 18. Appl Environ Microbiol. 2008. PMID: 18424547 Free PMC article.
-
Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli.Appl Environ Microbiol. 2010 Apr;76(8):2397-401. doi: 10.1128/AEM.02902-09. Epub 2010 Feb 12. Appl Environ Microbiol. 2010. PMID: 20154114 Free PMC article.
-
Escherichia coli strains engineered for homofermentative production of D-lactic acid from glycerol.Appl Environ Microbiol. 2010 Jul;76(13):4327-36. doi: 10.1128/AEM.00664-10. Epub 2010 May 14. Appl Environ Microbiol. 2010. PMID: 20472739 Free PMC article.
-
Anaerobic respiration in engineered Escherichia coli with an internal electron acceptor to produce fuel ethanol.Ann N Y Acad Sci. 2008 Mar;1125:363-72. doi: 10.1196/annals.1419.020. Ann N Y Acad Sci. 2008. PMID: 18378606 Review.
-
Development of ethanologenic bacteria.Adv Biochem Eng Biotechnol. 2007;108:237-61. doi: 10.1007/10_2007_068. Adv Biochem Eng Biotechnol. 2007. PMID: 17665158 Review.
Cited by
-
Efficient synthesis of L-lactic acid from glycerol by metabolically engineered Escherichia coli.Microb Cell Fact. 2013 Jan 25;12:7. doi: 10.1186/1475-2859-12-7. Microb Cell Fact. 2013. PMID: 23347598 Free PMC article.
-
Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant.Appl Environ Microbiol. 2010 Oct;76(19):6591-9. doi: 10.1128/AEM.01484-10. Epub 2010 Aug 6. Appl Environ Microbiol. 2010. PMID: 20693441 Free PMC article.
-
Redesigning Escherichia coli metabolism for anaerobic production of isobutanol.Appl Environ Microbiol. 2011 Jul;77(14):4894-904. doi: 10.1128/AEM.00382-11. Epub 2011 Jun 3. Appl Environ Microbiol. 2011. PMID: 21642415 Free PMC article.
-
In silico profiling of Escherichia coli and Saccharomyces cerevisiae as terpenoid factories.Microb Cell Fact. 2013 Sep 23;12:84. doi: 10.1186/1475-2859-12-84. Microb Cell Fact. 2013. PMID: 24059635 Free PMC article.
-
Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post-transfer refinement.Elife. 2014 Nov 24;3:e04279. doi: 10.7554/eLife.04279. Elife. 2014. PMID: 25418043 Free PMC article.
References
-
- Barbirato, F., C. Camarasa-Claret, J. Grivet, and A. Bories. 1995. Glycerol fermentation by a new 1,3-propanediol-producing microorganism: Enterobacter agglomerans. Appl. Microbiol. Biotechnol. 43:786-793.
-
- Biebl, H. 1991. Glycerol fermentation of 1,3-propanediol by Clostridium butyricum. Measurement of product inhibition by use of a pH-auxostat. Appl. Microbiol. Biotechnol. 35:701-705.
-
- Boenigk, R., S. Bowien, and G. Gottschalk. 1993. Fermentation of glycerol to 1,3-propanediol in continuous cultures of Citrobacter freundii. Appl. Microbiol. Biotechnol. 38:453-457.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

