SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo
- PMID: 19734346
- PMCID: PMC2770661
- DOI: 10.1093/nar/gkp685
SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo
Abstract
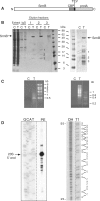
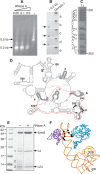
DEAD-box proteins play specific roles in remodeling RNA or ribonucleoprotein complexes. Yet, in vitro, they generally behave as nonspecific RNA-dependent ATPases, raising the question of what determines their specificity in vivo. SrmB, one of the five Escherichia coli DEAD-box proteins, participates in the assembly of the large ribosomal subunit. Moreover, when overexpressed, it compensates for a mutation in L24, the ribosomal protein (r-protein) thought to initiate assembly. Here, using the tandem affinity purification (TAP) procedure, we show that SrmB forms a complex with r-proteins L4, L24 and a region near the 5'-end of 23S rRNA that binds these proteins. In vitro reconstitution experiments show that the stability of this complex reflects cooperative interactions of SrmB with L4, L24 and rRNA. These observations are consistent with an early role of SrmB in assembly and explain the genetic link between SrmB and L24. Besides its catalytic core, SrmB possesses a nonconserved C-terminal extension that, we show, is not essential for SrmB function and specificity. In this regard, SrmB differs from DbpA, another DEAD-box protein involved in ribosome assembly.
Figures
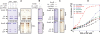
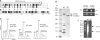
Similar articles
-
SrmB Rescues Trapped Ribosome Assembly Intermediates.J Mol Biol. 2020 Feb 14;432(4):978-990. doi: 10.1016/j.jmb.2019.12.013. Epub 2019 Dec 23. J Mol Biol. 2020. PMID: 31877323 Free PMC article.
-
Identification of the sites of action of SrmB, a DEAD-box RNA helicase involved in Escherichia coli ribosome assembly.Mol Microbiol. 2011 Oct;82(2):300-11. doi: 10.1111/j.1365-2958.2011.07779.x. Epub 2011 Aug 22. Mol Microbiol. 2011. PMID: 21859437
-
Structural basis for the activation of the DEAD-box RNA helicase DbpA by the nascent ribosome.Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2105961118. doi: 10.1073/pnas.2105961118. Proc Natl Acad Sci U S A. 2021. PMID: 34453003 Free PMC article.
-
Assembly of bacterial ribosomes.Annu Rev Biochem. 2011;80:501-26. doi: 10.1146/annurev-biochem-062608-160432. Annu Rev Biochem. 2011. PMID: 21529161 Review.
-
Functions of DEAD-box proteins in bacteria: current knowledge and pending questions.Biochim Biophys Acta. 2013 Aug;1829(8):866-77. doi: 10.1016/j.bbagrm.2013.01.012. Epub 2013 Feb 13. Biochim Biophys Acta. 2013. PMID: 23415794 Review.
Cited by
-
A Listeria monocytogenes RNA helicase essential for growth and ribosomal maturation at low temperatures uses its C terminus for appropriate interaction with the ribosome.J Bacteriol. 2012 Aug;194(16):4377-85. doi: 10.1128/JB.00348-12. Epub 2012 Jun 15. J Bacteriol. 2012. PMID: 22707705 Free PMC article.
-
DEAD Box RNA Helicases: Biochemical Properties, Role in RNA Processing and Ribosome Biogenesis.Cell Biochem Biophys. 2024 Jun;82(2):427-434. doi: 10.1007/s12013-024-01240-w. Epub 2024 Mar 2. Cell Biochem Biophys. 2024. PMID: 38430409 Review.
-
Long-term ecological and evolutionary dynamics in the gut microbiomes of carbapenemase-producing Enterobacteriaceae colonized subjects.Nat Microbiol. 2022 Oct;7(10):1516-1524. doi: 10.1038/s41564-022-01221-w. Epub 2022 Sep 15. Nat Microbiol. 2022. PMID: 36109646 Free PMC article.
-
Fluorescence bimolecular complementation enables facile detection of ribosome assembly defects in Escherichia coli.RNA Biol. 2016 Sep;13(9):872-82. doi: 10.1080/15476286.2016.1207037. Epub 2016 Jul 7. RNA Biol. 2016. PMID: 27388791 Free PMC article.
-
SrmB Rescues Trapped Ribosome Assembly Intermediates.J Mol Biol. 2020 Feb 14;432(4):978-990. doi: 10.1016/j.jmb.2019.12.013. Epub 2019 Dec 23. J Mol Biol. 2020. PMID: 31877323 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

