Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors
- PMID: 19734888
- PMCID: PMC2910619
- DOI: 10.1038/ncb1970
Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors
Abstract
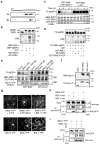
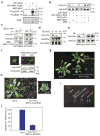
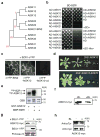
Brassinosteroid (BR) regulates gene expression and plant development through a receptor kinase-mediated signal transduction pathway. Despite the identification of many components of this pathway, it remains unclear how the BR signal is transduced from the cell surface to the nucleus. Here we describe a complete BR signalling pathway by elucidating key missing steps. We show that phosphorylation of BSK1 (BR-signalling kinase 1) by the BR receptor kinase BRI1 (BR-insensitive 1) promotes BSK1 binding to the BSU1 (BRI1 suppressor 1) phosphatase, and BSU1 inactivates the GSK3-like kinase BIN2 (BR-insensitive 2) by dephosphorylating a conserved phospho-tyrosine residue (pTyr 200). Mutations that affect phosphorylation/dephosphorylation of BIN2 pTyr200 (bin2-1, bin2-Y200F and quadruple loss-of-function of BSU1-related phosphatases) support an essential role for BSU1-mediated BIN2 dephosphorylation in BR-dependent plant growth. These results demonstrate direct sequential BR activation of BRI1, BSK1 and BSU1, and inactivation of BIN2, leading to accumulation of unphosphorylated BZR (brassinazole resistant) transcription factors in the nucleus. This study establishes a fully connected BR signalling pathway and provides new insights into the mechanism of GSK3 regulation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. - PubMed
-
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. - PubMed
-
- Belkhadir Y, Wang X, Chory J. Arabidopsis brassinosteroid signaling pathway. Sci STKE. 2006;2006:cm5. - PubMed
-
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

