Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation
- PMID: 19734905
- PMCID: PMC2751862
- DOI: 10.1038/ni.1783
Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation
Erratum in
- Nat Immunol. 2010 Jan;11(1):97
Abstract
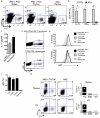
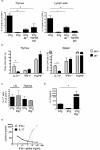
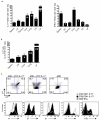
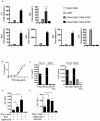
Interleukin 17 (IL-17)-producing CD4(+) helper T cells (T(H)-17 cells) share a developmental relationship with Foxp3(+) regulatory T cells (T(reg) cells). Here we show that a T(H)-17 population differentiates in the thymus in a manner influenced by recognition of self antigen and by the cytokines IL-6 and transforming growth factor-beta (TGF-beta). Like previously described T(H)-17 cells, the T(H)-17 cells that developed in the thymus expressed the transcription factor RORgamma t and the IL-23 receptor. These cells also expressed alpha(4)beta(1) integrins and the chemokine receptor CCR6 and were recruited to the lung, gut and liver. In the liver, these cells secreted IL-22 in response to self antigen and mediated host protection during inflammation. Thus, T(H)-17 cells, like T(reg) cells, can be selected by self antigens in the thymus.
Figures



Comment in
-
The importance of being earnestly selfish.Nat Immunol. 2009 Oct;10(10):1047-9. doi: 10.1038/ni1009-1047. Nat Immunol. 2009. PMID: 19767722 Free PMC article.
Similar articles
-
Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells.Nat Immunol. 2007 Sep;8(9):942-9. doi: 10.1038/ni1496. Epub 2007 Aug 5. Nat Immunol. 2007. PMID: 17676045
-
IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways.Nat Immunol. 2007 Sep;8(9):967-74. doi: 10.1038/ni1488. Epub 2007 Jun 20. Nat Immunol. 2007. PMID: 17581537
-
Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells.Nat Immunol. 2009 Aug;10(8):864-71. doi: 10.1038/ni.1770. Epub 2009 Jul 5. Nat Immunol. 2009. PMID: 19578368
-
Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection.Curr Top Microbiol Immunol. 2005;293:43-71. doi: 10.1007/3-540-27702-1_3. Curr Top Microbiol Immunol. 2005. PMID: 15981475 Review.
-
IL-17 and Th17 Cells.Annu Rev Immunol. 2009;27:485-517. doi: 10.1146/annurev.immunol.021908.132710. Annu Rev Immunol. 2009. PMID: 19132915 Review.
Cited by
-
Selection of self-reactive T cells in the thymus.Annu Rev Immunol. 2012;30:95-114. doi: 10.1146/annurev-immunol-020711-075035. Epub 2011 Dec 5. Annu Rev Immunol. 2012. PMID: 22149933 Free PMC article. Review.
-
Myelin basic protein-specific TCR/HLA-DRB5*01:01 transgenic mice support the etiologic role of DRB5*01:01 in multiple sclerosis.J Immunol. 2012 Sep 15;189(6):2897-908. doi: 10.4049/jimmunol.1103087. Epub 2012 Aug 10. J Immunol. 2012. PMID: 22888134 Free PMC article.
-
T-cell receptor affinity in thymic development.Immunology. 2012 Apr;135(4):261-7. doi: 10.1111/j.1365-2567.2011.03547.x. Immunology. 2012. PMID: 22182461 Free PMC article. Review.
-
Adaptive immune responses to Candida albicans infection.Virulence. 2015;6(4):327-37. doi: 10.1080/21505594.2015.1004977. Epub 2015 Jan 21. Virulence. 2015. PMID: 25607781 Free PMC article. Review.
-
Diversity of IL-17-producing T lymphocytes.Cell Mol Life Sci. 2013 Jul;70(13):2271-90. doi: 10.1007/s00018-012-1163-6. Epub 2012 Oct 4. Cell Mol Life Sci. 2013. PMID: 23052209 Free PMC article. Review.
References
-
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. - PubMed
-
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. - PubMed
-
- Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. - PubMed
-
- Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. - PubMed
-
- Ivanov, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

