Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly
- PMID: 19734909
- PMCID: PMC2759411
- DOI: 10.1038/nm.2023
Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly
Abstract
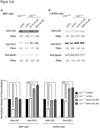
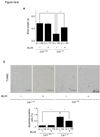
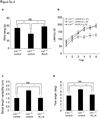
Lissencephaly is a devastating neurological disorder caused by defective neuronal migration. LIS1 (official symbol PAFAH1B1, for platelet-activating factor acetylhydrolase, isoform 1b, subunit 1) was identified as the gene mutated in individuals with lissencephaly, and it was found to regulate cytoplasmic dynein function and localization. Here we show that inhibition or knockdown of calpains protects LIS1 from proteolysis, resulting in the augmentation of LIS1 amounts in Lis1(+/-) mouse embryonic fibroblast cells and rescue of the aberrant distribution of cytoplasmic dynein, mitochondria and beta-COP-positive vesicles. We also show that calpain inhibitors improve neuronal migration of Lis1(+/-) cerebellar granular neurons. Intraperitoneal injection of the calpain inhibitor ALLN to pregnant Lis1(+/-) dams rescued apoptotic neuronal cell death and neuronal migration defects in Lis1(+/-) offspring. Furthermore, in utero knockdown of calpain by short hairpin RNA rescued defective cortical layering in Lis1(+/-) mice. Thus, calpain inhibition is a potential therapeutic intervention for lissencephaly.
Figures






Similar articles
-
ALLN rescues an in vitro excitatory synaptic transmission deficit in Lis1 mutant mice.J Neurophysiol. 2013 Jan;109(2):429-36. doi: 10.1152/jn.00431.2012. Epub 2012 Oct 24. J Neurophysiol. 2013. PMID: 23100132 Free PMC article.
-
[Molecular mechanism of lissencephaly--how LIS1 and NDEL1 regulate cytoplasmic dynein?].Brain Nerve. 2008 Apr;60(4):375-81. Brain Nerve. 2008. PMID: 18421979 Review. Japanese.
-
Genetic enhancement of the Lis1+/- phenotype by a heterozygous mutation in the adenomatous polyposis coli gene.Dev Neurosci. 2008;30(1-3):157-70. doi: 10.1159/000109860. Dev Neurosci. 2008. PMID: 18075263 Free PMC article.
-
Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice.J Neurosci. 2009 Dec 9;29(49):15520-30. doi: 10.1523/JNEUROSCI.4630-09.2009. J Neurosci. 2009. PMID: 20007476 Free PMC article.
-
LIS1 functions in normal development and disease.Curr Opin Neurobiol. 2013 Dec;23(6):951-6. doi: 10.1016/j.conb.2013.08.001. Epub 2013 Aug 23. Curr Opin Neurobiol. 2013. PMID: 23973156 Review.
Cited by
-
A Gastrointestinal Calpain Complex, G-calpain, Is a Heterodimer of CAPN8 and CAPN9 Calpain Isoforms, Which Play Catalytic and Regulatory Roles, Respectively.J Biol Chem. 2016 Dec 30;291(53):27313-27322. doi: 10.1074/jbc.M116.763912. Epub 2016 Nov 23. J Biol Chem. 2016. PMID: 27881674 Free PMC article.
-
Alpha-synuclein facilitates to form short unconventional microtubules that have a unique function in the axonal transport.Sci Rep. 2017 Nov 27;7(1):16386. doi: 10.1038/s41598-017-15575-3. Sci Rep. 2017. PMID: 29180624 Free PMC article.
-
Calpain 8/nCL-2 and calpain 9/nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense.PLoS Genet. 2010 Jul 29;6(7):e1001040. doi: 10.1371/journal.pgen.1001040. PLoS Genet. 2010. PMID: 20686710 Free PMC article.
-
Neurodevelopmental Genetic Diseases Associated With Microdeletions and Microduplications of Chromosome 17p13.3.Front Genet. 2018 Mar 23;9:80. doi: 10.3389/fgene.2018.00080. eCollection 2018. Front Genet. 2018. PMID: 29628935 Free PMC article. Review.
-
Developmental downregulation of LIS1 expression limits axonal extension and allows axon pruning.Biol Open. 2017 Jul 15;6(7):1041-1055. doi: 10.1242/bio.025999. Biol Open. 2017. PMID: 28630356 Free PMC article.
References
-
- Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. - PubMed
-
- Gleeson JG, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. - PubMed
-
- Pilz DT, et al. LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum Mol Genet. 1998;7:2029–2037. - PubMed
-
- Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. - PubMed
-
- Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected] Nature. 1994;370:216–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

