The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye
- PMID: 19736327
- PMCID: PMC2739146
- DOI: 10.1242/dev.038422
The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye
Abstract
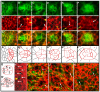
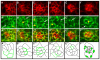
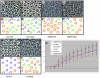
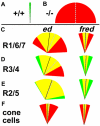
Directed cellular movements are a universal feature of morphogenesis in multicellular organisms. Differential adhesion between the stationary and motile cells promotes these cellular movements to effect spatial patterning of cells. A prominent feature of Drosophila eye development is the 90 degrees rotational movement of the multicellular ommatidial precursors within a matrix of stationary cells. We demonstrate that the cell adhesion molecules Echinoid (Ed) and Friend of Echinoid (Fred) act throughout ommatidial rotation to modulate the degree of ommatidial precursor movement. We propose that differential levels of Ed and Fred between stationary and rotating cells at the initiation of rotation create a permissive environment for cell movement, and that uniform levels in these two populations later contribute to stopping the movement. Based on genetic data, we propose that ed and fred impart a second, independent, ;brake-like' contribution to this process via Egfr signaling. Ed and Fred are localized in largely distinct and dynamic patterns throughout rotation. However, ed and fred are required in only a subset of cells - photoreceptors R1, R7 and R6 - for normal rotation, cells that have only recently been linked to a role in planar cell polarity (PCP). This work also provides the first demonstration of a requirement for cone cells in the ommatidial rotation aspect of PCP. ed and fred also genetically interact with the PCP genes, but affect only the degree-of-rotation aspect of the PCP phenotype. Significantly, we demonstrate that at least one PCP protein, Stbm, is required in R7 to control the degree of ommatidial rotation.
Figures



References
-
- Bai, J., Chiu, W., Wang, J., Tzeng, T., Perrimon, N. and Hsu, J. (2001). The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development 128, 591-601. - PubMed
-
- Bastock, R., Strutt, H. and Strutt, D. (2003). Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development 130, 3007-3014. - PubMed
-
- Brown, K. E. and Freeman, M. (2003). Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development 130, 5401-5412. - PubMed
-
- Chae, J., Kim, M. J., Goo, J. H., Collier, S., Gubb, D., Charlton, J., Adler, P. N. and Park, W. J. (1999). The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126, 5421-5429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

