Panorama phylogenetic diversity and distribution of type A influenza viruses based on their six internal gene sequences
- PMID: 19737421
- PMCID: PMC2746212
- DOI: 10.1186/1743-422X-6-137
Panorama phylogenetic diversity and distribution of type A influenza viruses based on their six internal gene sequences
Abstract
Background: Type A influenza viruses are important pathogens of humans, birds, pigs, horses and some marine mammals. The viruses have evolved into multiple complicated subtypes, lineages and sublineages. Recently, the phylogenetic diversity of type A influenza viruses from a whole view has been described based on the viral external HA and NA gene sequences, but remains unclear in terms of their six internal genes (PB2, PB1, PA, NP, MP and NS).
Methods: In this report, 2798 representative sequences of the six viral internal genes were selected from GenBank using the web servers in NCBI Influenza Virus Resource. Then, the phylogenetic relationships among the representative sequences were calculated using the software tools MEGA 4.1 and RAxML 7.0.4. Lineages and sublineages were classified mainly according to topology of the phylogenetic trees and distribution of the viruses in hosts, regions and time.
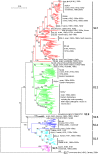
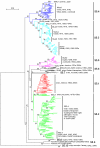
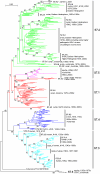
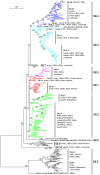
Results: The panorama phylogenetic trees of the six internal genes of type A influenza viruses were constructed. Lineages and sublineages within the type based on the six internal genes were classified and designated by a tentative universal numerical nomenclature system. The diversity of influenza viruses circulating in different regions, periods, and hosts based on the panorama trees was analyzed.
Conclusion: This study presents the first whole views to the phylogenetic diversity and distribution of type A influenza viruses based on their six internal genes. It also proposes a tentative universal nomenclature system for the viral lineages and sublineages. These can be a candidate framework to generalize the history and explore the future of the viruses, and will facilitate future scientific communications on the phylogenetic diversity and evolution of the viruses. In addition, it provides a novel phylogenetic view (i.e. the whole view) to recognize the viruses including the origin of the pandemic A(H1N1) influenza viruses.
Figures


References
-
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel type A influenza virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

