Translational informatics: enabling high-throughput research paradigms
- PMID: 19737991
- PMCID: PMC2789669
- DOI: 10.1152/physiolgenomics.00050.2009
Translational informatics: enabling high-throughput research paradigms
Abstract
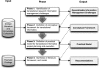
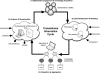
A common thread throughout the clinical and translational research domains is the need to collect, manage, integrate, analyze, and disseminate large-scale, heterogeneous biomedical data sets. However, well-established and broadly adopted theoretical and practical frameworks and models intended to address such needs are conspicuously absent in the published literature or other reputable knowledge sources. Instead, the development and execution of multidisciplinary, clinical, or translational studies are significantly limited by the propagation of "silos" of both data and expertise. Motivated by this fundamental challenge, we report upon the current state and evolution of biomedical informatics as it pertains to the conduct of high-throughput clinical and translational research and will present both a conceptual and practical framework for the design and execution of informatics-enabled studies. The objective of presenting such findings and constructs is to provide the clinical and translational research community with a common frame of reference for discussing and expanding upon such models and methodologies.
Figures


References
-
- Aksit M, Kurtev I. Elsevier special issue on foundations and applications of model driven architecture. Science Computer Programming 73: 1–2, 2008
-
- Ardekani AM, Akhondi MM, Sadeghi MR. Application of genomic and proteomic technologies to early detection of cancer. Archives Iranian Medicine 11: 427–434, 2008 - PubMed
-
- Ariyachandra T, Watson HJ. Which data warehouse architecture is best? Commun ACM 51: 146–147, 2008
-
- Batra D. Unified modeling language (UML) topics: the past, the problems, and the prospects. J Database Management 19: I–VII, 2008
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

