Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells
- PMID: 19740739
- PMCID: PMC2781530
- DOI: 10.1074/jbc.M109.016162
Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells
Abstract
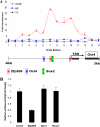
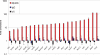
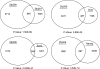
Zfp206 (recently renamed Zscan10) encodes a zinc finger transcription factor specifically expressed in human and mouse embryonic stem cells (ESC). It has been shown that Zfp206 is required to maintain ESC in an undifferentiated, pluripotent state. Presented here are data showing that Zfp206 works together with two other transcription factors, Oct4 and Sox2, which are also essential regulators of ESC pluripotency. We show that Zfp206 binds to the Oct4 promoter and directly regulates Oct4 expression. Genome-wide mapping of Zfp206-binding sites in ESC identifies more than 3000 target genes, many of which encode transcription factors that are also targeted for regulation by Oct4 and Sox2. In addition, we show that Zfp206 physically interacts with both Oct4 and Sox2. These data demonstrate that Zfp206 is a key component of the core transcriptional regulatory network and together with Oct4 and Sox2 regulates differentiation of ESC.
Figures




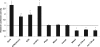
References
-
- Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) Nat. Genet. 38, 431–440 - PubMed
-
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006) Nature 442, 533–538 - PubMed
-
- Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 - PubMed
-
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Cell 133, 1106–1117 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

