Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein
- PMID: 19740757
- PMCID: PMC2781561
- DOI: 10.1074/jbc.M109.044800
Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein
Abstract
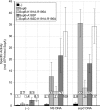
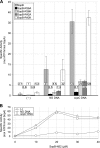
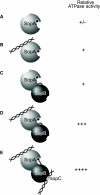
In bacteria, mitotic stability of plasmids and many chromosomes depends on replicon-specific systems, which comprise a centromere, a centromere-binding protein and an ATPase. Dynamic self-assembly of the ATPase appears to enable active partition of replicon copies into cell-halves, but for Walker-box partition ATPases the molecular mechanism is unknown. ATPase activity appears to be essential for this process. DNA and centromere-binding proteins are known to stimulate the ATPase activity but molecular details of the stimulation mechanism have not been reported. We have investigated the interactions which stimulate ATP hydrolysis by the SopA partition ATPase of plasmid F. By using SopA and SopB proteins deficient in DNA binding, we have found that the intrinsic ability of SopA to hydrolyze ATP requires direct DNA binding by SopA but not by SopB. Our results show that two independent interactions of SopA act in synergy to stimulate its ATPase. SopA must interact with (i) DNA, through its ATP-dependent nonspecific DNA binding domain and (ii) SopB, which we show here to provide an arginine-finger motif. In addition, the latter interaction stimulates ATPase maximally when SopB is part of the partition complex. Hence, our data demonstrate that DNA acts on SopA in two ways, directly as nonspecific DNA and through SopB as centromeric DNA, to fully activate SopA ATP hydrolysis.
Figures


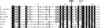

Similar articles
-
Polymerization of SopA partition ATPase: regulation by DNA binding and SopB.Mol Microbiol. 2007 Jan;63(2):468-81. doi: 10.1111/j.1365-2958.2006.05537.x. Epub 2006 Dec 11. Mol Microbiol. 2007. PMID: 17166176
-
Defining the role of ATP hydrolysis in mitotic segregation of bacterial plasmids.PLoS Genet. 2013;9(12):e1003956. doi: 10.1371/journal.pgen.1003956. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24367270 Free PMC article.
-
Role of the ATP-binding site of SopA protein in partition of the F plasmid.J Mol Biol. 2001 Nov 30;314(3):387-99. doi: 10.1006/jmbi.2001.5158. J Mol Biol. 2001. PMID: 11846553
-
Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation.Biochem J. 2008 May 15;412(1):1-18. doi: 10.1042/BJ20080359. Biochem J. 2008. PMID: 18426389 Review.
-
Surfing biological surfaces: exploiting the nucleoid for partition and transport in bacteria.Mol Microbiol. 2012 Nov;86(3):513-23. doi: 10.1111/mmi.12017. Epub 2012 Sep 19. Mol Microbiol. 2012. PMID: 22934804 Free PMC article. Review.
Cited by
-
The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.Nat Commun. 2021 Aug 27;12(1):5166. doi: 10.1038/s41467-021-25429-2. Nat Commun. 2021. PMID: 34453062 Free PMC article.
-
CTP and parS coordinate ParB partition complex dynamics and ParA-ATPase activation for ParABS-mediated DNA partitioning.Elife. 2021 Jul 21;10:e65651. doi: 10.7554/eLife.65651. Elife. 2021. PMID: 34286695 Free PMC article.
-
PomX, a ParA/MinD ATPase activating protein, is a triple regulator of cell division in Myxococcus xanthus.Elife. 2021 Mar 18;10:e66160. doi: 10.7554/eLife.66160. Elife. 2021. PMID: 33734087 Free PMC article.
-
Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):E1390-7. doi: 10.1073/pnas.1302745110. Epub 2013 Mar 11. Proc Natl Acad Sci U S A. 2013. PMID: 23479605 Free PMC article.
-
Protein gradients on the nucleoid position the carbon-fixing organelles of cyanobacteria.Elife. 2018 Dec 6;7:e39723. doi: 10.7554/eLife.39723. Elife. 2018. PMID: 30520729 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

