Tryptophan residues in the portal protein of herpes simplex virus 1 critical to the interaction with scaffold proteins and incorporation of the portal into capsids
- PMID: 19740984
- PMCID: PMC2772705
- DOI: 10.1128/JVI.01463-09
Tryptophan residues in the portal protein of herpes simplex virus 1 critical to the interaction with scaffold proteins and incorporation of the portal into capsids
Abstract
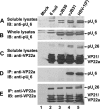
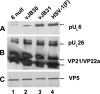
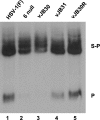
Incorporation of the herpes simplex virus 1 (HSV-1) portal vertex into the capsid requires interaction with a 12-amino-acid hydrophobic domain within capsid scaffold proteins. The goal of this work was to identify domains and residues in the UL6-encoded portal protein pUL6 critical to the interaction with scaffold proteins. We show that whereas the wild-type portal and scaffold proteins readily coimmunoprecipitated with one another in the absence of other viral proteins, truncation beyond the first 18 or last 36 amino acids of the portal protein precluded this coimmunoprecipitation. The coimmunoprecipitation was also precluded by mutation of conserved tryptophan (W) residues to alanine (A) at positions 27, 90, 127, 163, 241, 262, 532, and 596 of UL6. All of these W-to-A mutations precluded the rescue of a viral deletion mutant lacking UL6, except W163A, which supported replication poorly, and W596A, which fully rescued replication. A recombinant virus bearing the W596A mutation replicated and packaged DNA normally, and scaffold proteins readily coimmunoprecipitated with portal protein from lysates of infected cells. Thus, viral functions compensated for the W596A mutation's detrimental effects on the portal-scaffold interaction seen during transient expression of portal and scaffold proteins. In contrast, the W27A mutation precluded portal-scaffold interactions in infected cell lysates, reduced the solubility of pUL6, decreased incorporation of the portal into capsids, and abrogated viral-DNA cleavage and packaging.
Figures




References
-
- Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. - PubMed
-
- Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex type 1. J. Gen. Virol. 73:2709-2713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

