Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly
- PMID: 19740989
- PMCID: PMC2772684
- DOI: 10.1128/JVI.01508-09
Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly
Abstract
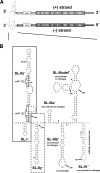
Hepatitis C virus (HCV) is a positive-strand RNA virus replicating its genome via a negative-strand [(-)] intermediate. Little is known about replication signals residing in the 3' end of HCV (-) RNA. Recent studies identified seven stem-loop structures (SL-I', -IIz', -IIy', -IIIa', -IIIb', -IIIcdef', and -IV') in this region. In the present study, we mapped the minimal region required for RNA replication to SL-I' and -IIz', functionally confirmed the SL-IIz' structure, and identified SL-IIIa' to -IV' as auxiliary replication elements. In addition, we show that the 5' nontranslated region of the genome most likely does not contain cis-acting RNA structures required for RNA packaging into infectious virions.
Figures



References
-
- Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. - PubMed
-
- Dutkiewicz, M., A. Swiatkowska, M. Figlerowicz, and J. Ciesiolka. 2008. Structural domains of the 3′-terminal sequence of the hepatitis C virus replicative strand. Biochemistry 47:12197-12207. - PubMed
-
- Fan, Z., Q. R. Yang, J. S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

