Genomic views of distant-acting enhancers
- PMID: 19741700
- PMCID: PMC2923221
- DOI: 10.1038/nature08451
Genomic views of distant-acting enhancers
Abstract
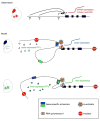
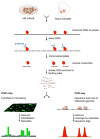
In contrast to protein-coding sequences, the significance of variation in non-coding DNA in human disease has been minimally explored. A great number of recent genome-wide association studies suggest that non-coding variation is a significant risk factor for common disorders, but the mechanisms by which this variation contributes to disease remain largely obscure. Distant-acting transcriptional enhancers--a major category of functional non-coding DNA--are involved in many developmental and disease-relevant processes. Genome-wide approaches to their discovery and functional characterization are now available and provide a growing knowledge base for the systematic exploration of their role in human biology and disease susceptibility.
Figures

References
-
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420 (6915):520–562. - PubMed
-
- Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316 (5830):1491–1493. - PubMed
-
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. Comprehensive overview of functional classes of gene regulatory sequences, including many disease-relevant examples identified through gene-centric studies. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

