The dimeric proto-ribosome: Structural details and possible implications on the origin of life
- PMID: 19742176
- PMCID: PMC2738903
- DOI: 10.3390/ijms10072921
The dimeric proto-ribosome: Structural details and possible implications on the origin of life
Abstract
A symmetric pocket-like entity, composed of two L-shaped RNA units, encircles the peptide synthesis site within the contemporary ribosome. This entity was suggested to be the vestige of a dimeric proto-ribosome, which could have formed spontaneously in the prebiotic world, catalyzing non-coded peptide bond formation and elongation. This structural element, beyond offering the initial step in the evolution of translation, is hypothesized here to be linked to the origin of life. By catalyzing the production of random peptide chains, the proto-ribosome could have enabled the formation of primary enzymes, launching a process of co-evolution of the translation apparatus and the proteins, thus presenting an alternative to the RNA world hypothesis.
Keywords: origin of life; proto-ribosome; ribosome symmetrical region.
Figures

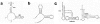
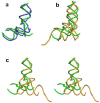
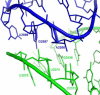
References
-
- Agmon I, Bashan A, Yonath A. On ribosome conservation and evolution. Israel Journal of Ecology and Evolution. 2006;52:359–374.
-
- Agmon I, Davidovich C, Bashan A, Yonath A. Identification of the prebiotic translation apparatus within the contemporary ribosome. 2009. http://precedings.nature.com/documents/-2921/version/1, accessed 18 March 2009.
-
- Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen HA, Harms J, Kessler M, Peretz M, Schluenzen F, Yonath A, Zarivach R. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Eur. J. Biochem. 2003;270:2543–2556. - PubMed
-
- Agmon I, Bashan A, Zarivach R, Yonath A. Symmetry at the active site of the ribosome: Structural and functional implications. Biol. Chem. 2005;386:833–844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

