Intestinal microbiota regulate xenobiotic metabolism in the liver
- PMID: 19742318
- PMCID: PMC2734986
- DOI: 10.1371/journal.pone.0006958
Intestinal microbiota regulate xenobiotic metabolism in the liver
Abstract
Background: The liver is the central organ for xenobiotic metabolism (XM) and is regulated by nuclear receptors such as CAR and PXR, which control the metabolism of drugs. Here we report that gut microbiota influences liver gene expression and alters xenobiotic metabolism in animals exposed to barbiturates.
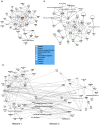
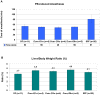
Principal findings: By comparing hepatic gene expression on microarrays from germfree (GF) and conventionally-raised mice (SPF), we identified a cluster of 112 differentially expressed target genes predominantly connected to xenobiotic metabolism and pathways inhibiting RXR function. These findings were functionally validated by exposing GF and SPF mice to pentobarbital which confirmed that xenobiotic metabolism in GF mice is significantly more efficient (shorter time of anesthesia) when compared to the SPF group.
Conclusion: Our data demonstrate that gut microbiota modulates hepatic gene expression and function by altering its xenobiotic response to drugs without direct contact with the liver.
Conflict of interest statement
Figures


References
-
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. - PubMed
-
- Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. - PubMed
-
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

