Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins
- PMID: 19744324
- PMCID: PMC2746813
- DOI: 10.1186/1471-2148-9-229
Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins
Abstract
Background: The co-translational incorporation of selenocysteine into nascent polypeptides by recoding the UGA stop codon occurs in all domains of life. In eukaryotes, this event requires at least three specific factors: SECIS binding protein 2 (SBP2), a specific translation elongation factor (eEFSec), selenocysteinyl tRNA, and a cis-acting selenocysteine insertion sequence (SECIS) element in selenoprotein mRNAs. While the phylogenetic relationships of selenoprotein families and the evolution of selenocysteine usage are well documented, the evolutionary history of SECIS binding proteins has not been explored.
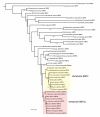
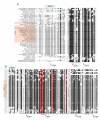
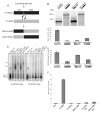
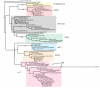
Results: In this report we present a phylogeny of the eukaryotic SECIS binding protein family which includes SBP2 and a related protein we herein term SBP2L. Here we show that SBP2L is an SBP2 paralogue in vertebrates and is the only form of SECIS binding protein in invertebrate deuterostomes, suggesting a key role in Sec incorporation in these organisms, but an SBP2/SBP2L fusion protein is unable to support Sec incorporation in vitro. An in-depth phylogenetic analysis of the conserved L7Ae RNA binding domain suggests an ancestral relationship with ribosomal protein L30. In addition, we describe the emergence of a motif upstream of the SBP2 RNA binding domain that shares significant similarity with a motif within the pseudouridine synthase Cbf5.
Conclusion: Our analysis suggests that SECIS binding proteins arose once in evolution but diverged significantly in multiple lineages. In addition, likely due to a gene duplication event in the early vertebrate lineage, SBP2 and SBP2L are paralogous in vertebrates.
Figures





Similar articles
-
Selenocysteine insertion sequence binding protein 2L is implicated as a novel post-transcriptional regulator of selenoprotein expression.PLoS One. 2012;7(4):e35581. doi: 10.1371/journal.pone.0035581. Epub 2012 Apr 17. PLoS One. 2012. PMID: 22530054 Free PMC article.
-
Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2.RNA Biol. 2014;11(11):1402-13. doi: 10.1080/15476286.2014.996472. RNA Biol. 2014. PMID: 25692238 Free PMC article.
-
Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes.Nat Struct Mol Biol. 2005 May;12(5):408-16. doi: 10.1038/nsmb922. Epub 2005 Apr 10. Nat Struct Mol Biol. 2005. PMID: 15821744
-
Protein factors mediating selenoprotein synthesis.Curr Protein Pept Sci. 2002 Feb;3(1):143-51. doi: 10.2174/1389203023380783. Curr Protein Pept Sci. 2002. PMID: 12370018 Review.
-
On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins.Biochim Biophys Acta Gen Subj. 2018 Nov;1862(11):2463-2472. doi: 10.1016/j.bbagen.2018.03.018. Epub 2018 Mar 17. Biochim Biophys Acta Gen Subj. 2018. PMID: 29555379 Free PMC article. Review.
Cited by
-
Differential co-expression network analysis elucidated genes associated with sensitivity to farnesyltransferase inhibitor and prognosis of acute myeloid leukemia.Cancer Med. 2023 Dec;12(24):22420-22436. doi: 10.1002/cam4.6804. Epub 2023 Dec 8. Cancer Med. 2023. PMID: 38069522 Free PMC article.
-
Synthesis and decoding of selenocysteine and human health.Croat Med J. 2012 Dec;53(6):535-50. doi: 10.3325/cmj.2012.53.535. Croat Med J. 2012. PMID: 23275319 Free PMC article. Review.
-
Lokiarchaeota Marks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems.Mol Biol Evol. 2016 Sep;33(9):2441-53. doi: 10.1093/molbev/msw122. Epub 2016 Jul 12. Mol Biol Evol. 2016. PMID: 27413050 Free PMC article.
-
The expression of essential selenoproteins during development requires SECIS-binding protein 2-like.Life Sci Alliance. 2022 Feb 24;5(5):e202101291. doi: 10.26508/lsa.202101291. Print 2022 May. Life Sci Alliance. 2022. PMID: 35210313 Free PMC article.
-
SECISBP2L-Mediated Selenoprotein Synthesis Is Essential for Autonomous Regulation of Oligodendrocyte Differentiation.J Neurosci. 2022 Jul 27;42(30):5860-5869. doi: 10.1523/JNEUROSCI.2141-21.2022. Epub 2022 Jun 27. J Neurosci. 2022. PMID: 35760530 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

