Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins
- PMID: 19745225
- PMCID: PMC2781523
- DOI: 10.1074/jbc.M109.041830
Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins
Abstract
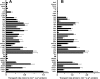
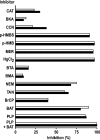
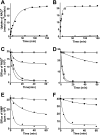
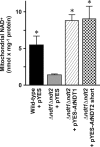
The Arabidopsis thaliana L. genome contains 58 membrane proteins belonging to the mitochondrial carrier family. Two mitochondrial carrier family members, here named AtNDT1 and AtNDT2, exhibit high structural similarities to the mitochondrial nicotinamide adenine dinucleotide (NAD(+)) carrier ScNDT1 from bakers' yeast. Expression of AtNDT1 or AtNDT2 restores mitochondrial NAD(+) transport activity in a yeast mutant lacking ScNDT. Localization studies with green fluorescent protein fusion proteins provided evidence that AtNDT1 resides in chloroplasts, whereas only AtNDT2 locates to mitochondria. Heterologous expression in Escherichia coli followed by purification, reconstitution in proteoliposomes, and uptake experiments revealed that both carriers exhibit a submillimolar affinity for NAD(+) and transport this compound in a counter-exchange mode. Among various substrates ADP and AMP are the most efficient counter-exchange substrates for NAD(+). Atndt1- and Atndt2-promoter-GUS plants demonstrate that both genes are strongly expressed in developing tissues and in particular in highly metabolically active cells. The presence of both carriers is discussed with respect to the subcellular localization of de novo NAD(+) biosynthesis in plants and with respect to both the NAD(+)-dependent metabolic pathways and the redox balance of chloroplasts and mitochondria.
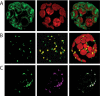
Figures


References
-
- Heldt H. W. (2005) Plant Biochemistry, Elsevier Academic Press, Burlington, MA
-
- Roux S. J., Steinebrunner I. (2007) Trends Plant Sci. 12, 522–527 - PubMed
-
- Lehninger A. L., Nelson D. L., Cox M. M. (1994) Prinzipien der Biochemie, Spektrum, Akad. Verlag, Heidelberg, Berlin, Oxford
-
- Klingenberg M. (2008) Biochim. Biophys. Acta 1778, 1978–2021 - PubMed
-
- Strotmann H., Berger S. (1969) Biochem. Biophys. Res. Com. 35, 20–26 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

