A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation
- PMID: 19745810
- PMCID: PMC2771092
- DOI: 10.1038/emboj.2009.256
A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation
Abstract
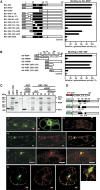
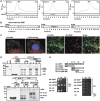
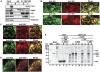
The plant viral re-initiation factor transactivator viroplasmin (TAV) activates translation of polycistronic mRNA by a re-initiation mechanism involving translation initiation factor 3 (eIF3) and the 60S ribosomal subunit (60S). QJ;Here, we report a new plant factor-re-initiation supporting protein (RISP)-that enhances TAV function in re-initiation. RISP interacts physically with TAV in vitro and in vivo. Mutants defective in interaction are less active, or inactive, in transactivation and viral amplification. RISP alone can serve as a scaffold protein, which is able to interact with eIF3 subunits a/c and 60S, apparently through the C-terminus of ribosomal protein L24. RISP pre-bound to eIF3 binds 40S, suggesting that RISP enters the translational machinery at the 43S formation step. RISP, TAV and 60S co-localize in epidermal cells of infected plants, and eIF3-TAV-RISP-L24 complex formation can be shown in vitro. These results suggest that RISP and TAV bridge interactions between eIF3-bound 40S and L24 of 60S after translation termination to ensure 60S recruitment during repetitive initiation events on polycistronic mRNA; RISP can thus be considered as a new component of the cell translation machinery.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Phosphorylation of a reinitiation supporting protein, RISP, determines its function in translation reinitiation.Nucleic Acids Res. 2021 Jul 9;49(12):6908-6924. doi: 10.1093/nar/gkab501. Nucleic Acids Res. 2021. PMID: 34133725 Free PMC article.
-
Control of translation reinitiation on the cauliflower mosaic virus (CaMV) polycistronic RNA.Biochem Soc Trans. 2004 Aug;32(Pt 4):592-6. doi: 10.1042/BST0320592. Biochem Soc Trans. 2004. PMID: 15270684
-
A plant viral "reinitiation" factor interacts with the host translational machinery.Cell. 2001 Sep 21;106(6):723-33. doi: 10.1016/s0092-8674(01)00487-1. Cell. 2001. PMID: 11572778
-
Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation.EMBO Rep. 2009 May;10(5):459-65. doi: 10.1038/embor.2009.70. Epub 2009 Apr 17. EMBO Rep. 2009. PMID: 19373251 Free PMC article. Review.
-
eIF3: a versatile scaffold for translation initiation complexes.Trends Biochem Sci. 2006 Oct;31(10):553-62. doi: 10.1016/j.tibs.2006.08.005. Epub 2006 Aug 22. Trends Biochem Sci. 2006. PMID: 16920360 Review.
Cited by
-
Dynamic changes in ribosome-associated proteome and phosphoproteome during deoxynivalenol-induced translation inhibition and ribotoxic stress.Toxicol Sci. 2014 Mar;138(1):217-33. doi: 10.1093/toxsci/kft270. Epub 2013 Nov 27. Toxicol Sci. 2014. PMID: 24284785 Free PMC article.
-
Setting Up Shop: The Formation and Function of the Viral Factories of Cauliflower mosaic virus.Front Plant Sci. 2017 Oct 30;8:1832. doi: 10.3389/fpls.2017.01832. eCollection 2017. Front Plant Sci. 2017. PMID: 29163571 Free PMC article. Review.
-
Auxin Signaling in Regulation of Plant Translation Reinitiation.Front Plant Sci. 2017 Jun 14;8:1014. doi: 10.3389/fpls.2017.01014. eCollection 2017. Front Plant Sci. 2017. PMID: 28659957 Free PMC article. Review.
-
Translational Regulation of Cytoplasmic mRNAs.Arabidopsis Book. 2013 Jul 18;11:e0165. doi: 10.1199/tab.0165. Print 2013. Arabidopsis Book. 2013. PMID: 23908601 Free PMC article.
-
Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses.Viruses. 2023 Mar 23;15(4):816. doi: 10.3390/v15040816. Viruses. 2023. PMID: 37112798 Free PMC article. Review.
References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 - PubMed
-
- Bonneville J-M, Sanfaçon H, Fütterer J, Hohn T (1989) Posttranscriptional transactivation in cauliflower mosaic virus. Cell 59: 1135–1143 - PubMed
-
- Bureau M, Leh V, Haas M, Geldreich A, Ryabova L, Yot P, Keller M (2004) P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J Gen Virol 85: 3765–3775 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

