Threading the needle: getting selenocysteine into proteins
- PMID: 19747061
- PMCID: PMC2864665
- DOI: 10.1089/ars.2009.2878
Threading the needle: getting selenocysteine into proteins
Abstract
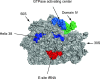
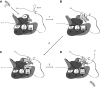
The co-translational incorporation of selenocysteine (Sec) requires that UGA be recognized as a sense rather than a nonsense codon. This is accomplished by the concerted action of a Sec insertion sequence (SECIS) element, SECIS binding protein 2, and a ternary complex of the Sec specific elongation factor, Sec-tRNA(Sec), and GTP. The mechanism by which they alter the canonical protein synthesis reaction has been elusive. Here we present an overview of the mechanistic perspective on Sec incorporation, highlighting recent advances in the field.
Figures
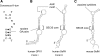

References
-
- Alsina B. Corominas M. Berry MJ. Baguña J. Serras F. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 1999;112:2875–2884. - PubMed
-
- Andersen GR. Pedersen L. Valente L. Chatterjee I. Kinzy TG. Kjeldgaard M. Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol Cell. 2000;6:1261–1266. - PubMed
-
- Boulon S. Marmier–Gourrier N. Pradet–Balade B. Wurth L. Verheggen C. Jády BE. Rothé B. Pescia C. Robert MC. Kiss T. Bardoni B. Krol A. Branlant C. Allmang C. Bertrand E. Charpentier B. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

