Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA
- PMID: 19748360
- PMCID: PMC2771690
- DOI: 10.1016/j.molcel.2009.07.013
Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA
Abstract
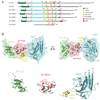
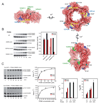
Siz1 is a founding member of the Siz/PIAS RING family of SUMO E3 ligases. The X-ray structure of an active Siz1 ligase revealed an elongated tripartite architecture comprised of an N-terminal PINIT domain, a central zinc-containing RING-like SP-RING domain, and a C-terminal domain we term the SP-CTD. Structure-based mutational analysis and biochemical studies show that the SP-RING and SP-CTD are required for activation of the E2 approximately SUMO thioester, while the PINIT domain is essential for redirecting SUMO conjugation to the proliferating cell nuclear antigen (PCNA) at lysine 164, a nonconsensus lysine residue that is not modified by the SUMO E2 in the absence of Siz1. Mutational analysis of Siz1 and PCNA revealed surfaces on both proteins that are required for efficient SUMO modification of PCNA in vitro and in vivo.
Figures





References
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. - PubMed
-
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. - PubMed
-
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

