Evidence that human Chlamydia pneumoniae was zoonotically acquired
- PMID: 19749045
- PMCID: PMC2786552
- DOI: 10.1128/JB.00746-09
Evidence that human Chlamydia pneumoniae was zoonotically acquired
Abstract
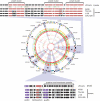
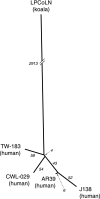
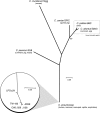
Zoonotic infections are a growing threat to global health. Chlamydia pneumoniae is a major human pathogen that is widespread in human populations, causing acute respiratory disease, and has been associated with chronic disease. C. pneumoniae was first identified solely in human populations; however, its host range now includes other mammals, marsupials, amphibians, and reptiles. Australian koalas (Phascolarctos cinereus) are widely infected with two species of Chlamydia, C. pecorum and C. pneumoniae. Transmission of C. pneumoniae between animals and humans has not been reported; however, two other chlamydial species, C. psittaci and C. abortus, are known zoonotic pathogens. We have sequenced the 1,241,024-bp chromosome and a 7.5-kb cryptic chlamydial plasmid of the koala strain of C. pneumoniae (LPCoLN) using the whole-genome shotgun method. Comparative genomic analysis, including pseudogene and single-nucleotide polymorphism (SNP) distribution, and phylogenetic analysis of conserved genes and SNPs against the human isolates of C. pneumoniae show that the LPCoLN isolate is basal to human isolates. Thus, we propose based on compelling genomic and phylogenetic evidence that humans were originally infected zoonotically by an animal isolate(s) of C. pneumoniae which adapted to humans primarily through the processes of gene decay and plasmid loss, to the point where the animal reservoir is no longer required for transmission.
Figures


References
-
- Balin, B. J., C. S. Little, C. J. Hammond, D. M. Appelt, J. A. Whittum-Hudson, H. C. Gerard, and A. P. Hudson. 2008. Chlamydophila pneumoniae and the etiology of late-onset Alzheimer's disease. J. Alzheimers Dis. 13:371-380. - PubMed
-
- Bodetti, T. J., E. Jacobson, C. Wan, L. Hafner, A. Pospischil, K. Rose, and P. Timms. 2002. Molecular evidence to support the expansion of the host range of Chlamydophila pneumoniae to include reptiles as well as humans, horses, koalas and amphibians. Syst. Appl. Microbiol. 25:146-152. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

