Archaeal intrinsic transcription termination in vivo
- PMID: 19749050
- PMCID: PMC2772485
- DOI: 10.1128/JB.00982-09
Archaeal intrinsic transcription termination in vivo
Abstract
Thermococcus kodakarensis (formerly Thermococcus kodakaraensis) strains have been constructed with synthetic and natural DNA sequences, predicted to function as archaeal transcription terminators, identically positioned between a constitutive promoter and a beta-glycosidase-encoding reporter gene (TK1761). Expression of the reporter gene was almost fully inhibited by the upstream presence of 5'-TTTTTTTT (T(8)) and was reduced >70% by archaeal intergenic sequences that contained oligo(T) sequences. An archaeal intergenic sequence (t(mcrA)) that conforms to the bacterial intrinsic terminator motif reduced TK1761 expression approximately 90%, but this required only the oligo(T) trail sequence and not the inverted-repeat and loop region. Template DNAs were amplified from each T. kodakarensis strain, and transcription in vitro by T. kodakarensis RNA polymerase was terminated by sequences that reduced TK1761 expression in vivo. Termination occurred at additional sites on these linear templates, including at a 5'-AAAAAAAA (A(8)) sequence that did not reduce TK1761 expression in vivo. When these sequences were transcribed on supercoiled plasmid templates, termination occurred almost exclusively at oligo(T) sequences. The results provide the first in vivo experimental evidence for intrinsic termination of archaeal transcription and confirm that archaeal transcription termination is stimulated by oligo(T) sequences and is different from the RNA hairpin-dependent mechanism established for intrinsic bacterial termination.
Figures
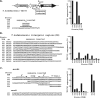

 ), ribosome binding site (RBS), and 5′ sequence of TK1761 (shaded box) are identified (34, 35). The additional sequences present in templates 552, 553, 554, and 557 are listed with the T8, A8, and (TA)4 sequences boxed. (B) Electrophoretic separation of the transcripts synthesized in vitro from linear (upper gel) and supercoiled, plasmid (lower gel) templates. The linear templates were generated by PCR as for panel A. The plasmid templates were the plasmids used to transform T. kodakarensis KW128 to generate T. kodakarensis strains TS372, TS552, TS553, TS554, and TS557. The [32P]ApC-labeled transcripts were separated by electrophoresis, visualized, and quantified using a Storm 840 phosphorimager (GE Healthcare). Transcripts that were terminated from +10 on templates 372 and 557 are identified above and below the upper gel, respectively, by the nucleotides at that position in the 372 and 557 sequences (see panel A). The transcripts that were terminated within T8, A8, and (TA)4 on the linear templates are boxed. The regions of the gels containing abortive, terminated, runoff (↓), or read-through (
), ribosome binding site (RBS), and 5′ sequence of TK1761 (shaded box) are identified (34, 35). The additional sequences present in templates 552, 553, 554, and 557 are listed with the T8, A8, and (TA)4 sequences boxed. (B) Electrophoretic separation of the transcripts synthesized in vitro from linear (upper gel) and supercoiled, plasmid (lower gel) templates. The linear templates were generated by PCR as for panel A. The plasmid templates were the plasmids used to transform T. kodakarensis KW128 to generate T. kodakarensis strains TS372, TS552, TS553, TS554, and TS557. The [32P]ApC-labeled transcripts were separated by electrophoresis, visualized, and quantified using a Storm 840 phosphorimager (GE Healthcare). Transcripts that were terminated from +10 on templates 372 and 557 are identified above and below the upper gel, respectively, by the nucleotides at that position in the 372 and 557 sequences (see panel A). The transcripts that were terminated within T8, A8, and (TA)4 on the linear templates are boxed. The regions of the gels containing abortive, terminated, runoff (↓), or read-through ( ) transcripts are indicated. (C) Percentages of transcripts initiated and extended beyond +10 that were terminated on linear versus circular versions of each template.
) transcripts are indicated. (C) Percentages of transcripts initiated and extended beyond +10 that were terminated on linear versus circular versions of each template.
 ,
,  ) (B) transcripts are indicated. (C) Percentages of transcripts initiated and extended beyond +10 that were terminated on linear versus circular versions of each template.
) (B) transcripts are indicated. (C) Percentages of transcripts initiated and extended beyond +10 that were terminated on linear versus circular versions of each template.References
-
- Bartlett, M. S. 2005. Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 8:677-684. - PubMed
-
- Braglia, P., R. Percudani, and G. Dieci. 2005. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 280:19551-19562. - PubMed
-
- Cavicchioli, R. (ed.). 2007. Archaea. Molecular and cellular biology. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

