A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association
- PMID: 19749154
- PMCID: PMC2768941
- DOI: 10.1105/tpc.108.063420
A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association
Abstract
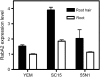
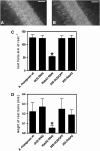
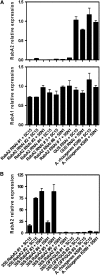
Legume plants are able to establish a symbiotic relationship with soil bacteria from the genus Rhizobium, leading to the formation of nitrogen-fixing root nodules. Successful nodulation requires both the formation of infection threads (ITs) in the root epidermis and the activation of cell division in the cortex to form the nodule primordium. This study describes the characterization of RabA2, a common bean (Phaseolus vulgaris) cDNA previously isolated as differentially expressed in root hairs infected with Rhizobium etli, which encodes a protein highly similar to small GTPases of the RabA2 subfamily. This gene is expressed in roots, particularly in root hairs, where the protein was found to be associated with vesicles that move along the cell. The role of this gene during nodulation has been studied in common bean transgenic roots using a reverse genetic approach. Examination of root morphology in RabA2 RNA interference (RNAi) plants revealed that the number and length of the root hairs were severely reduced in these plants. Upon inoculation with R. etli, nodulation was completely impaired and no induction of early nodulation genes (ENODs), such as ERN1, ENOD40, and Hap5, was detected in silenced hairy roots. Moreover, RabA2 RNAi plants failed to induce root hair deformation and to initiate ITs, indicating that morphological changes that precede bacterial infection are compromised in these plants. We propose that RabA2 acts in polar growth of root hairs and is required for reorientation of the root hair growth axis during bacterial infection.
Figures





References
-
- Benedito, V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55 504–513. - PubMed
-
- Bibikova, T.N., Blancaflor, E.B., and Gilroy, S. (1999). Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17 657–665. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

