Identification of gemin5 as a novel 7-methylguanosine cap-binding protein
- PMID: 19750007
- PMCID: PMC2736588
- DOI: 10.1371/journal.pone.0007030
Identification of gemin5 as a novel 7-methylguanosine cap-binding protein
Abstract
Background: A unique attribute of RNA molecules synthesized by RNA polymerase II is the presence of a 7-methylguanosine (m(7)G) cap structure added co-transcriptionally to the 5' end. Through its association with trans-acting effector proteins, the m(7)G cap participates in multiple aspects of RNA metabolism including localization, translation and decay. However, at present relatively few eukaryotic proteins have been identified as factors capable of direct association with m(7)G.
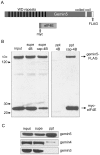
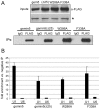
Methodology/principal findings: Employing an unbiased proteomic approach, we identified gemin5, a component of the survival of motor neuron (SMN) complex, as a factor capable of direct and specific interaction with the m(7)G cap. Gemin5 was readily purified by cap-affinity chromatography in contrast to other SMN complex proteins. Investigating the underlying basis for this observation, we found that purified gemin5 associates with m(7)G-linked sepharose in the absence of detectable eIF4E, and specifically crosslinks to radiolabeled cap structure after UV irradiation. Deletion analysis revealed that an intact set of WD repeat domains located in the N-terminal half of gemin5 are required for cap-binding. Moreover, using structural modeling and site-directed mutagenesis, we identified two proximal aromatic residues located within the WD repeat region that significantly impact m(7)G association.
Conclusions/significance: This study rigorously identifies gemin5 as a novel cap-binding protein and describes an unprecedented role for WD repeat domains in m(7)G recognition. The findings presented here will facilitate understanding of gemin5's role in the metabolism of non-coding snRNAs and perhaps other RNA pol II transcripts.
Conflict of interest statement
Figures





References
-
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. - PubMed
-
- Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J Biol Chem. 1999;274:16553–16562. - PubMed
-
- Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. - PubMed
-
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. - PubMed
-
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

