Global and local mobility of apocalmodulin monitored through fast-field cycling relaxometry
- PMID: 19751682
- PMCID: PMC2749786
- DOI: 10.1016/j.bpj.2009.07.005
Global and local mobility of apocalmodulin monitored through fast-field cycling relaxometry
Abstract
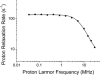
Calmodulin (CaM) is a ubiquitous eukaryotic protein with two conformationally independent domains that can bind up to two calcium ions each. In the calcium-bound state, CaM is able to regulate a vast number of cellular activities by binding to a multiplicity of target proteins in different modes. Its versatility has been ascribed to its anomalously high flexibility. The calcium-free form (apoCaM), which is the resting state of CaM in cells, is also able to functionally bind a number of protein targets, but its dynamics has received less attention. At variance with the calcium-bound form, the crystal structure of apoCaM shows a compact organization of the two domains, but NMR measurements could not detect any contact between them, thus indicating the presence of mobility in solution. The mobility of apoCaM is here investigated through protein proton relaxation rate measurements performed with a high-sensitivity fast-field cycling relaxometer. Such measurements provide direct access to the spectral density function and show that 1), the reorientation time is in agreement with a closed form of the protein; but 2), the collective order parameter is much smaller than for other well folded compact proteins, indicating that a remarkably large side-chain mobility must be present.
Figures

References
-
- Eisenmesser E.Z., Bosco D.A., Akke M., Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. - PubMed
-
- Lindorff-Larsen K., Best R.B., DePristo M.A., Dobson C.M., Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. - PubMed
-
- Fragai M., Luchinat C., Parigi G. “Four-dimensional” protein structures: examples from metalloproteins. Acc. Chem. Res. 2006;39:909–917. - PubMed
-
- Lange O.F., Lakomek N.-A., Farès C., Schröder G.F., Walter K.F.A. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

