Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion
- PMID: 19752025
- PMCID: PMC2753158
- DOI: 10.1083/jcb.200906098
Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion
Abstract
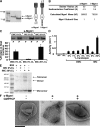
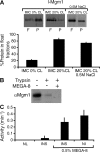
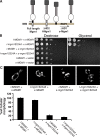
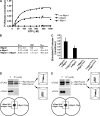
Two dynamin-related protein (DRP) families are essential for fusion of the outer and inner mitochondrial membranes, Fzo1 (yeast)/Mfn1/Mfn2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), respectively. Fzo1/Mfns possess two medial transmembrane domains, which place their critical GTPase and coiled-coil domains in the cytosol. In contrast, Mgm1/Opa1 are present in cells as long (l) isoforms that are anchored via the N terminus to the inner membrane, and short (s) isoforms were predicted to be soluble in the intermembrane space. We addressed the roles of Mgm1 isoforms and how DRPs function in membrane fusion. Our analysis indicates that in the absence of a membrane, l- and s-Mgm1 both exist as inactive GTPase monomers, but that together in trans they form a functional dimer in a cardiolipin-dependent manner that is the building block for higher-order assemblies.
Figures

References
-
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling.Cell. 126:163–175 doi:10.1016/j.cell.2006.06.021 - DOI - PubMed
-
- Crowther R.A., Henderson R., Smith J.M. 1996. MRC image processing programs.J. Struct. Biol. 116:9–16 doi:10.1006/jsbi.1996.0003 - DOI - PubMed
-
- Danino D., Hinshaw J.E. 2001. Dynamin family of mechanoenzymes.Curr. Opin. Cell Biol. 13:454–460 doi:10.1016/S0955-0674(00)00236-2 - DOI - PubMed
-
- Daum G., Vance J.E. 1997. Import of lipids into mitochondria.Prog. Lipid Res. 36:103–130 doi:10.1016/S0163-7827(97)00006-4 - DOI - PubMed
-
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N., et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol Chem.J. Biol. Chem. 281:37972–37979 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

