Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1
- PMID: 19752031
- PMCID: PMC2786451
- DOI: 10.1128/IAI.00551-09
Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1
Abstract
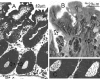
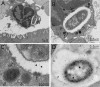
Currently, the Golden Syrian hamster is widely considered an important model of Clostridium difficile disease, as oral infection of this animal pretreated with antibiotics reproduces many of the symptoms observed in humans. Two C. difficile strains, B1 and 630, showed significant differences in the progression and severity of disease in this model. B1-infected hamsters exhibited more severe pathology and a shorter time to death than hamsters infected with 630. Histological changes in the gut did not correlate with absolute numbers of C. difficile bacteria, but there were clear differences in the distribution of bacteria within gut tissues. Light, scanning, and transmission electron microscopy revealed high numbers of B1 bacteria at the mucosal surface of the tissue, whereas 630 bacteria were more frequently associated with the crypt regions. Both B1 and 630 bacteria were frequently observed within polymorphonuclear leukocytes, although, interestingly, a space frequently separated B1 bacteria from the phagosome wall, a phenomenon not observed with 630. However, pilus-like structures were detected on 630 located in the crypts of the gut tissue. Furthermore, B1 bacteria, but not 630 bacteria, were found within nonphagocytic cells, including enterocytes and muscle cells.
Figures






References
-
- Barbut, F., B. Gariazzo, L. Bonne, V. Lalande, B. Burghoffer, R. Luiuz, and J.-C. Petit. 2007. Clinical features of Clostridium difficile—associated infections and molecular characterisation of strains: results of a restrospective study, 2000-2004. Infect. Control Hosp. Epidemiol. 28:131-139. - PubMed
-
- Bartlett, J. G., A. B. Onderdonk, R. L. Cisneros, and D. L. Kasper. 1977. Clindamycin associated colitis due to a toxin producing species of Clostridium in hamsters. J. Infect. Dis. 136:701-705. - PubMed
-
- Borriello, S. P., J. M. Ketley, T. M. Mitchell, F. E. Barclay, A. R. Welch, A. B. Price, and J. Stephen. 1987. Clostridium difficile—a spectrum of virulence and analysis of putative virulence determinants in the hamster model of antibiotic-associated colitis. J. Med. Microbiol. 24:53-64. - PubMed
-
- Cerquetti, M., A. Serafino, A. Sebastianelli, and P. Mastrantonio. 2002. Binding of Clostridium difficile to Caco-2 epithelial cell line and extracellular matrix proteins. FEMS Immunol. Med. Microbiol. 32:211-218. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

