A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis
- PMID: 19754970
- PMCID: PMC2759933
- DOI: 10.1186/1752-0509-3-92
A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis
Abstract
Background: Because metabolism is fundamental in sustaining microbial life, drugs that target pathogen-specific metabolic enzymes and pathways can be very effective. In particular, the metabolic challenges faced by intracellular pathogens, such as Mycobacterium tuberculosis, residing in the infected host provide novel opportunities for therapeutic intervention.
Results: We developed a mathematical framework to simulate the effects on the growth of a pathogen when enzymes in its metabolic pathways are inhibited. Combining detailed models of enzyme kinetics, a complete metabolic network description as modeled by flux balance analysis, and a dynamic cell population growth model, we quantitatively modeled and predicted the dose-response of the 3-nitropropionate inhibitor on the growth of M. tuberculosis in a medium whose carbon source was restricted to fatty acids, and that of the 5'-O-(N-salicylsulfamoyl) adenosine inhibitor in a medium with low-iron concentration.
Conclusion: The predicted results quantitatively reproduced the experimentally measured dose-response curves, ranging over three orders of magnitude in inhibitor concentration. Thus, by allowing for detailed specifications of the underlying enzymatic kinetics, metabolic reactions/constraints, and growth media, our model captured the essential chemical and biological factors that determine the effects of drug inhibition on in vitro growth of M. tuberculosis cells.
Figures

 , we analyzed the Metabolic Network to infer the biomass growth rate μ and substrate uptake rate vC. Using the Population Growth Model, we related biomass growth rate μ and substrate uptake rate vC to cell concentration [X]. We dynamically coupled the biomass growth rate and the diminished substrate concentration to develop a time-dependent model that dynamically infers cell concentration after the introduction of an inhibitor. Once these model components were specified, together with the initial substrate [C0] and cell [X0] concentrations in the growth medium, the calculations performed within this framework only required input in the form of a specific inhibitor concentration [I] to predict cellular growth.
, we analyzed the Metabolic Network to infer the biomass growth rate μ and substrate uptake rate vC. Using the Population Growth Model, we related biomass growth rate μ and substrate uptake rate vC to cell concentration [X]. We dynamically coupled the biomass growth rate and the diminished substrate concentration to develop a time-dependent model that dynamically infers cell concentration after the introduction of an inhibitor. Once these model components were specified, together with the initial substrate [C0] and cell [X0] concentrations in the growth medium, the calculations performed within this framework only required input in the form of a specific inhibitor concentration [I] to predict cellular growth.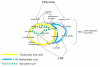


 ; (F) Michaelis-Menten rate constant KSUC,MCL1; and (G) Michaelis-Menten rate constant K3-NP,MCL1.
; (F) Michaelis-Menten rate constant KSUC,MCL1; and (G) Michaelis-Menten rate constant K3-NP,MCL1.


 ; (C) upper limit of glycerol uptake rate
; (C) upper limit of glycerol uptake rate  ; (D) upper limit of glycerol uptake rate
; (D) upper limit of glycerol uptake rate  ; and (E) time length of cellular growth t, which were each set to its original parameter value (solid line), the value increased by 50% (dashed line), and decreased by 50% (dotted line).
; and (E) time length of cellular growth t, which were each set to its original parameter value (solid line), the value increased by 50% (dashed line), and decreased by 50% (dotted line).References
-
- Joyce AR, Palsson BO. Toward whole cell modeling and simulation: comprehensive functional genomics through the constraint-based approach. Prog Drug Res. 2007;64:267–309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

