LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development
- PMID: 19755105
- PMCID: PMC2758028
- DOI: 10.1016/j.neuron.2009.07.031
LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development
Abstract
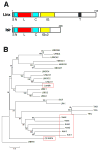
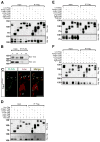
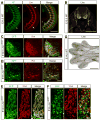
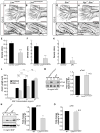
Genome-wide screens were performed to identify transmembrane proteins that mediate axonal growth, guidance and target field innervation of somatosensory neurons. One gene, Linx (alias Islr2), encoding a leucine-rich repeat and immunoglobulin (LIG) family protein, is expressed in a subset of developing sensory and motor neurons. Domain and genomic structures of Linx and other LIG family members suggest that they are evolutionarily related to Trk receptor tyrosine kinases (RTKs). Several LIGs, including Linx, are expressed in subsets of somatosensory and motor neurons, and select members interact with TrkA and Ret RTKs. Moreover, axonal projection defects in mice harboring a null mutation in Linx resemble those in mice lacking Ngf, TrkA, and Ret. In addition, Linx modulates NGF-TrkA- and GDNF-GFRalpha1/Ret-mediated axonal extension in cultured sensory and motor neurons, respectively. These findings show that LIGs physically interact with RTKs and modulate their activities to control axonal extension, guidance and branching.
Figures



Similar articles
-
GDNF and NGF family members and receptors in human fetal and adult spinal cord and dorsal root ganglia.J Comp Neurol. 2001 Nov 12;440(2):204-17. doi: 10.1002/cne.1380. J Comp Neurol. 2001. PMID: 11745618
-
IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life.Neuron. 1997 Oct;19(4):849-61. doi: 10.1016/s0896-6273(00)80966-6. Neuron. 1997. PMID: 9354331
-
Role of motoneuron-derived neurotrophin 3 in survival and axonal projection of sensory neurons during neural circuit formation.Development. 2012 Mar;139(6):1125-32. doi: 10.1242/dev.069997. Epub 2012 Feb 8. Development. 2012. PMID: 22318233
-
The GDNF family ligands and receptors - implications for neural development.Curr Opin Neurobiol. 2000 Feb;10(1):103-10. doi: 10.1016/s0959-4388(99)00048-3. Curr Opin Neurobiol. 2000. PMID: 10679429 Review.
-
NGF and ProNGF: Regulation of neuronal and neoplastic responses through receptor signaling.Adv Biol Regul. 2015 May;58:16-27. doi: 10.1016/j.jbior.2014.11.003. Epub 2014 Nov 20. Adv Biol Regul. 2015. PMID: 25491371 Free PMC article. Review.
Cited by
-
Usefulness of the measurement of neurite outgrowth of primary sensory neurons to study cancer-related painful complications.Biochem Pharmacol. 2021 Jun;188:114520. doi: 10.1016/j.bcp.2021.114520. Epub 2021 Mar 17. Biochem Pharmacol. 2021. PMID: 33741328 Free PMC article.
-
Neurotrophin signaling endosomes: biogenesis, regulation, and functions.Curr Opin Neurobiol. 2016 Aug;39:139-45. doi: 10.1016/j.conb.2016.06.004. Epub 2016 Jun 18. Curr Opin Neurobiol. 2016. PMID: 27327126 Free PMC article. Review.
-
Coronin-1 is a neurotrophin endosomal effector that is required for developmental competition for survival.Nat Neurosci. 2014 Jan;17(1):36-45. doi: 10.1038/nn.3593. Epub 2013 Nov 24. Nat Neurosci. 2014. PMID: 24270184 Free PMC article.
-
Uncoupling of molecular maturation from peripheral target innervation in nociceptors expressing a chimeric TrkA/TrkC receptor.PLoS Genet. 2014 Feb 6;10(2):e1004081. doi: 10.1371/journal.pgen.1004081. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24516396 Free PMC article.
-
Delineating neurotrophin-3 dependent signaling pathways underlying sympathetic axon growth along intermediate targets.Mol Cell Neurosci. 2017 Jul;82:66-75. doi: 10.1016/j.mcn.2017.04.011. Epub 2017 Apr 28. Mol Cell Neurosci. 2017. PMID: 28461220 Free PMC article.
References
-
- Airaksinen MS, Koltzenburg M, Lewin GR, Masu Y, Helbig C, Wolf E, Brem G, Toyka KV, Thoenen H, Meyer M. Specific subtypes of cutaneous mechanoreceptors require neurotrophin-3 following peripheral target innervation. Neuron. 1996;16:287–295. - PubMed
-
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. - PubMed
-
- Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. - PubMed
-
- Baloh RH, Enomoto H, Johnson EM, Jr, Milbrandt J. The GDNF family ligands and receptors - implications for neural development. Curr Opin Neurobiol. 2000;10:103–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

