Different strategies adopted by K(b) and L(d) to generate T cell specificity directed against their respective bound peptides
- PMID: 19755422
- PMCID: PMC2781669
- DOI: 10.1074/jbc.M109.040501
Different strategies adopted by K(b) and L(d) to generate T cell specificity directed against their respective bound peptides
Abstract
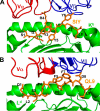
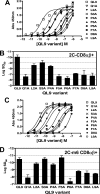
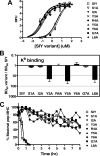
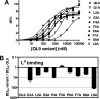
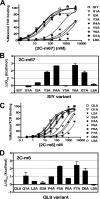
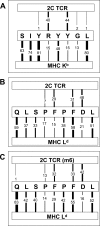
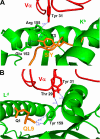
Mouse T cell clone 2C recognizes two different major histocompatibility (MHC) ligands, the self MHC K(b) and the allogeneic MHC L(d). Two distinct peptides, SIY (SIYRYYGL) and QL9 (QLSPFPFDL), act as strong and specific agonists when bound to K(b) and L(d), respectively. To explore further the mechanisms involved in peptide potency and specificity, here we examined a collection of single amino acid peptide variants of SIY and QL9 for 1) T cell activity, 2) binding to their respective MHC, and 3) binding to the 2C T cell receptor (TCR) and high affinity TCR mutants. Characterization of SIY binding to MHC K(b) revealed significant effects of three SIY residues that were clearly embedded within the K(b) molecule. In contrast, QL9 binding to MHC L(d) was influenced by the majority of peptide side chains, distributed across the entire length of the peptide. Binding of the SIY-K(b) complex to the TCR involved three SIY residues that were pointed toward the TCR, whereas again the majority of QL9 residues influenced binding of TCRs, and thus the QL9 residues had impacts on both L(d) and TCR binding. In general, the magnitude of T cell activity mediated by a peptide variant was influenced more by peptide binding to MHC than by binding the TCR, especially for higher affinity TCRs. Findings with both systems, but QL9-L(d) in particular, suggest that many single-residue substitutions, introduced into peptides to improve their binding to MHC and thus their vaccine potential, could impair T cell reactivity due to their dual impact on TCR binding.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

