The structural biology of type IV secretion systems
- PMID: 19756009
- PMCID: PMC3869563
- DOI: 10.1038/nrmicro2218
The structural biology of type IV secretion systems
Abstract
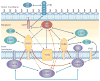
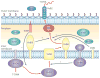
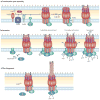
Type IV secretion systems (T4SSs) are versatile secretion systems that are found in both Gram-negative and Gram-positive bacteria and secrete a wide range of substrates, from single proteins to protein-protein and protein-DNA complexes. They usually consist of 12 components that are organized into ATP-powered, double-membrane-spanning complexes. The structures of single soluble components or domains have been solved, but an understanding of how these structures come together has only recently begun to emerge. This Review focuses on the structural advances that have been made over the past 10 years and how the corresponding structural insights have helped to elucidate many of the details of the mechanism of type IV secretion.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

