Efficient and rapid template-directed nucleic acid copying using 2'-amino-2',3'-dideoxyribonucleoside-5'-phosphorimidazolide monomers
- PMID: 19757789
- PMCID: PMC2759813
- DOI: 10.1021/ja906557v
Efficient and rapid template-directed nucleic acid copying using 2'-amino-2',3'-dideoxyribonucleoside-5'-phosphorimidazolide monomers
Abstract
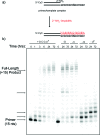
The development of a sequence-general nucleic acid copying system is an essential step in the assembly of a synthetic protocell, an autonomously replicating spatially localized chemical system capable of spontaneous Darwinian evolution. Previously described nonenzymatic template-copying experiments have validated the concept of nonenzymatic replication, but have not yet achieved robust, sequence-general polynucleotide replication. The 5'-phosphorimidazolides of the 2'-amino-2',3'-dideoxyribonucleotides are attractive as potential monomers for such a system because they polymerize by forming 2'-->5' linkages, which are favored in nonenzymatic polymerization reactions using similarly activated ribonucleotides on RNA templates. Furthermore, the 5'-activated 2'-amino nucleotides do not cyclize. We recently described the rapid and efficient nonenzymatic copying of a DNA homopolymer template (dC(15)) encapsulated within fatty acid vesicles using 2'-amino-2',3'-dideoxyguanosine-5'-phosphorimidazolide as the activated monomer. However, to realize a true Darwinian system, the template-copying chemistry must be able to copy most sequences and their complements to allow for the transmission of information from generation to generation. Here, we describe the copying of a series of nucleic acid templates using 2'-amino-2',3'-dideoxynucleotide-5'-phosphorimidazolides. Polymerization reactions proceed rapidly to completion on short homopolymer RNA and LNA templates, which favor an A-type duplex geometry. We show that more efficiently copied sequences are generated by replacing the adenine nucleobase with diaminopurine, and uracil with C5-(1-propynyl)uracil. Finally, we explore the copying of longer, mixed-sequence RNA templates to assess the sequence-general copying ability of 2'-amino-2',3'-dideoxynucleoside-5'-phosphorimidazolides. Our results are a significant step forward in the realization of a self-replicating genetic polymer compatible with protocell template copying and suggest that N2'-->P5'-phosphoramidate DNA may have the potential to function as a self-replicating system.
Figures







Similar articles
-
Nonenzymatic RNA-templated Synthesis of N3'→P5' Phosphoramidate DNA.Bio Protoc. 2020 Sep 5;10(17):e3734. doi: 10.21769/BioProtoc.3734. eCollection 2020 Sep 5. Bio Protoc. 2020. PMID: 33659395 Free PMC article.
-
Synthesis of N3'-P5'-linked phosphoramidate DNA by nonenzymatic template-directed primer extension.J Am Chem Soc. 2013 Jan 16;135(2):924-32. doi: 10.1021/ja311164j. Epub 2013 Jan 7. J Am Chem Soc. 2013. PMID: 23252395 Free PMC article.
-
Synthesis of activated 3'-amino-3'-deoxy-2-thio-thymidine, a superior substrate for the nonenzymatic copying of nucleic acid templates.Chem Commun (Camb). 2016 Mar 4;52(18):3684-6. doi: 10.1039/c5cc10317g. Chem Commun (Camb). 2016. PMID: 26857159
-
The Mechanism of Nonenzymatic Template Copying with Imidazole-Activated Nucleotides.Angew Chem Int Ed Engl. 2019 Aug 5;58(32):10812-10819. doi: 10.1002/anie.201902050. Epub 2019 Jul 4. Angew Chem Int Ed Engl. 2019. PMID: 30908802 Review.
-
The Emergence of RNA from the Heterogeneous Products of Prebiotic Nucleotide Synthesis.J Am Chem Soc. 2021 Mar 10;143(9):3267-3279. doi: 10.1021/jacs.0c12955. Epub 2021 Feb 26. J Am Chem Soc. 2021. PMID: 33636080 Review.
Cited by
-
Nonenzymatic Template-Directed Synthesis of Mixed-Sequence 3'-NP-DNA up to 25 Nucleotides Long Inside Model Protocells.J Am Chem Soc. 2019 Jul 3;141(26):10481-10488. doi: 10.1021/jacs.9b04858. Epub 2019 Jun 20. J Am Chem Soc. 2019. PMID: 31180644 Free PMC article.
-
Enzyme-free ligation of dimers and trimers to RNA primers.Nucleic Acids Res. 2019 May 7;47(8):3836-3845. doi: 10.1093/nar/gkz160. Nucleic Acids Res. 2019. PMID: 30869145 Free PMC article.
-
pH-controlled DNA- and RNA-templated assembly of short oligomers.Chem Sci. 2015 Jan 1;6(1):542-547. doi: 10.1039/c4sc03028a. Epub 2014 Oct 28. Chem Sci. 2015. PMID: 28936308 Free PMC article.
-
Four Ways to Oligonucleotides Without Phosphoimidazolides.J Mol Evol. 2016 Jan;82(1):5-10. doi: 10.1007/s00239-015-9709-5. Epub 2015 Oct 31. J Mol Evol. 2016. PMID: 26520151 Review.
-
Rewarming the Primordial Soup: Revisitations and Rediscoveries in Prebiotic Chemistry.Chembiochem. 2018 Jan 4;19(1):22-25. doi: 10.1002/cbic.201700534. Epub 2017 Nov 22. Chembiochem. 2018. PMID: 29164768 Free PMC article. Review.
References
-
- Szostak J. W.; Bartel D. P.; Luisi P. L. Nature 2001, 409, 387–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

