EnzyMiner: automatic identification of protein level mutations and their impact on target enzymes from PubMed abstracts
- PMID: 19758466
- PMCID: PMC2745584
- DOI: 10.1186/1471-2105-10-S8-S2
EnzyMiner: automatic identification of protein level mutations and their impact on target enzymes from PubMed abstracts
Abstract
Background: A better understanding of the mechanisms of an enzyme's functionality and stability, as well as knowledge and impact of mutations is crucial for researchers working with enzymes. Though, several of the enzymes' databases are currently available, scientific literature still remains at large for up-to-date source of learning the effects of a mutation on an enzyme. However, going through vast amounts of scientific documents to extract the information on desired mutation has always been a time consuming process. In this paper, therefore, we describe an unique method, termed as EnzyMiner, which automatically identifies the PubMed abstracts that contain information on the impact of a protein level mutation on the stability and/or the activity of a given enzyme.
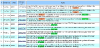
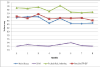
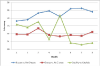
Results: We present an automated system which identifies the abstracts that contain an amino-acid-level mutation and then classifies them according to the mutation's effect on the enzyme. In the case of mutation identification, MuGeX, an automated mutation-gene extraction system has an accuracy of 93.1% with a 91.5 F-measure. For impact analysis, document classification is performed to identify the abstracts that contain a change in enzyme's stability or activity resulting from the mutation. The system was trained on lipases and tested on amylases with an accuracy of 85%.
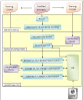
Conclusion: EnzyMiner identifies the abstracts that contain a protein mutation for a given enzyme and checks whether the abstract is related to a disease with the help of information extraction and machine learning techniques. For disease related abstracts, the mutation list and direct links to the abstracts are retrieved from the system and displayed on the Web. For those abstracts that are related to non-diseases, in addition to having the mutation list, the abstracts are also categorized into two groups. These two groups determine whether the mutation has an effect on the enzyme's stability or functionality followed by displaying these on the web.
Figures

References
-
- Renugopalakrishnan V, Garduno-Juarez R, Narasimhan G, Verma C, Wei X, Li P. Rational design of thermally stable proteins: relevance to bionanotechnology. J Nanosci Nanotechnol. 2005;5:1759–1767. - PubMed
-
- Hult K, Berglund P. Engineered enzymes for improved organic synthesis. Curr Opin Biotechnol. 2002;14:395–400. - PubMed
-
- Laskowski R, Wallace A. Enzyme Structures Database http://www.ebi.ac.uk/thornton-srv/databases/enzymes/
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

